Female Heterogamety in Madagascar Chameleons
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

An Intial Estimation of the Numbers and Identification of Extant Non
Answers Research Journal 8 (2015):171–186. www.answersingenesis.org/arj/v8/lizard-kinds-order-squamata.pdf $Q,QLWLDO(VWLPDWLRQRIWKH1XPEHUVDQG,GHQWLÀFDWLRQRI Extant Non-Snake/Non-Amphisbaenian Lizard Kinds: Order Squamata Tom Hennigan, Truett-McConnell College, Cleveland, Georgia. $EVWUDFW %LRV\VWHPDWLFVLVLQJUHDWÁX[WRGD\EHFDXVHRIWKHSOHWKRUDRIJHQHWLFUHVHDUFKZKLFKFRQWLQXDOO\ UHGHÀQHVKRZZHSHUFHLYHUHODWLRQVKLSVEHWZHHQRUJDQLVPV'HVSLWHWKHODUJHDPRXQWRIGDWDEHLQJ SXEOLVKHGWKHFKDOOHQJHLVKDYLQJHQRXJKNQRZOHGJHDERXWJHQHWLFVWRGUDZFRQFOXVLRQVUHJDUGLQJ WKHELRORJLFDOKLVWRU\RIRUJDQLVPVDQGWKHLUWD[RQRP\&RQVHTXHQWO\WKHELRV\VWHPDWLFVIRUPRVWWD[D LVLQJUHDWIOX[DQGQRWZLWKRXWFRQWURYHUV\E\SUDFWLWLRQHUVLQWKHILHOG7KHUHIRUHWKLVSUHOLPLQDU\SDSHU LVmeant to produce a current summary of lizard systematics, as it is understood today. It is meant to lay a JURXQGZRUNIRUFUHDWLRQV\VWHPDWLFVZLWKWKHJRDORIHVWLPDWLQJWKHQXPEHURIEDUDPLQVEURXJKWRQ WKH $UN %DVHG RQ WKH DQDO\VHV RI FXUUHQW PROHFXODU GDWD WD[RQRP\ K\EULGL]DWLRQ FDSDELOLW\ DQG VWDWLVWLFDO EDUDPLQRORJ\ RI H[WDQW RUJDQLVPV D WHQWDWLYH HVWLPDWH RI H[WDQW QRQVQDNH QRQ DPSKLVEDHQLDQOL]DUGNLQGVZHUHWDNHQRQERDUGWKH$UN,WLVKRSHGWKDWWKLVSDSHUZLOOHQFRXUDJH IXWXUHUHVHDUFKLQWRFUHDWLRQLVWELRV\VWHPDWLFV Keywords: $UN(QFRXQWHUELRV\VWHPDWLFVWD[RQRP\UHSWLOHVVTXDPDWDNLQGEDUDPLQRORJ\OL]DUG ,QWURGXFWLRQ today may change tomorrow, depending on the data Creation research is guided by God’s Word, which and assumptions about that data. For example, LVIRXQGDWLRQDOWRWKHVFLHQWLÀFPRGHOVWKDWDUHEXLOW naturalists assume randomness and universal 7KHELEOLFDODQGVFLHQWLÀFFKDOOHQJHLVWRLQYHVWLJDWH -

Implementation of Article 16, Council Regulation (EC) No. 338/97, in the EU
Implementation of Article 16, Council Regulation (EC) No. 338/97, in the 25 Member States of the European Union Tobias Garstecki Report commissioned by the European Commission Contract 07.0402/2005/399949/MAR/E2 Report prepared by TRAFFIC Europe for the European Commission in completion of Contract 07.0402/2005/399949/MAR/E2 All material appearing in this publication is copyrighted and may be reproduced with permission. Any reproduction in full or in part of this publication must credit the European Commission as the copyright owner. The views of the authors expressed in this publication do not necessarily reflect those of the European Commission or the TRAFFIC network, WWF or IUCN. The designation of geographical entities in this publication, and the presentation of the material, do not imply the expression of any opinion whatsoever on the part of the European Commission, TRAFFIC or its supporting organizations concerning the legal status of any country, territory, or area, or its authorities, or concerning the delimitation of its frontiers or boundaries. The TRAFFIC symbol copyright and Registered Trademark ownership is held by WWF. TRAFFIC is a joint programme of WWF and IUCN. Suggested citation: Garstecki, T (2006): Implementation of Article 16, Council Regulation (EC) No. 338/97, in the 25 Member States of the European Union. A TRAFFIC Europe Report for the European Commission, Brussels, Belgium. Implementation of Article 16, EC Regulation 338/97, in the 25 Member States of the European Union CONTENTS Acknowledgements 1 Executive Summary 2 Background and project description 3 Objectives 4 Comparison of penalties for wildlife trade regulation offences in EU Member States 4 Table 1: Comparison of minimum and maximum penalties and seizure/confiscation powers in relation to Article 16 of EC Regulation 338/97 in EU Member States 9 Procedures for determining penalties and monetary compensation 16 Overview 16 1. -

Genetic Relationship of Three Butterfly Lizard Species (Leiolepis Reevesii Rubritaeniata, Leiolepis Belliana Belliana, Leiolepis
Kasetsart J. (Nat. Sci.) 44 : 424 - 435 (2010) Genetic Relationship of Three Butterfly Lizard Species (Leiolepis reevesii rubritaeniata, Leiolepis belliana belliana, Leiolepis boehmei, Agamidae, Squamata) Inferred from Nuclear Gene Sequence Analyses Kornsorn Srikulnath1, 2, Kazumi Matsubara3, Yoshinobu Uno2, Amara Thongpan1, Saowanee Suputtitada1, Chizuko Nishida2, 3, Yoichi Matsuda2, 3, 4 and Somsak Apisitwanich1* ABSTRACT The genetic relationship was investigated of three butterfly lizard species (Leiolepis reevesii rubritaeniata, L. belliana belliana and L. boehmei) selectively inhabiting Thailand. The findings were based on RAG1 and C-mos gene analyses. The DNA sequences were also compared with the other squamate reptiles. The analysis strongly supported that L. reevesii rubritaeniata was related more closely to L. belliana belliana than to L. boehmei. The phylogenetic position of Leiolepis spp., however, was contentious with regard to its relationship among the Leiolepidinae, Agaminae and Chamaeleonidae, which suggested that their phylogeny remains uncertain. Keywords: butterfly lizard, Leiolepidinae, phylogeny, RAG1, C-mos INTRODUCTION inhabit Southeast Asia. They show a great variety of karyotypes and sexual systems. In Thailand, The Squamata is the most diverse there are three species, which barely can be reptilian order that has been classified traditionally discriminated from other congeneric species by into three suborders: Serpentes (snakes), their typical scale and skin coloration (Peters, Amphisbaenia (worm lizards) and Lacertilia 1971). Bisexualism has been described in Leiolepis (lizards). The extant lizards can be further belliana belliana (2n=2x=36), which is widely categorized into five infraorders (the Iguania, found throughout the country, L. belliana ocellata Gekkota, Scincomorpha, Diploglossa, Dibamia, (2n=2x=34) found in upper northern, and L. -

A List of the Herpetological Type Specimens in the Zoologisches Forschungsmuseum Alexander Koenig, Bonn
Bonn zoological Bulletin Volume 59 pp. 79–108 Bonn, December 2010 A list of the herpetological type specimens in the Zoologisches Forschungsmuseum Alexander Koenig, Bonn Wolfgang Böhme Zoologisches Forschungsmuseum Alexander Koenig, Herpetology Section, Adenauerallee 160, D-53113 Bonn, Germany; E-mail: [email protected]. Abstract. In the herpetological collection of ZFMK 528 scientific species group names are represented by type materi- al. Of these, 304 names are documented by primary type specimens (onomatophores) while for 224 further names sec- ondary type specimens (typoids) are available, ranging chronologically from 1801 to 2010. The list is a shortened pred- ecessor of a comprehensive type catalogue in progress. It lists name bearing types with their catalogue numbers includ- ing information on further type series members also in other institutions, while secondary types are listed only by pres- ence, both in ZFMK and other collections including holotype repositories. Geographic origin and currently valid names are also provided. Key words. Amphibians and reptiles, type list, ZFMK Bonn. INTRODUCTION A first ZFMK herpetological type catalogue was published (currently section) in 1951, for many decades. Nonethe- (Böhme 1974) three years after I had entered Museum less, the present list does comprise some historical “pre- Koenig as a herpetological curator. It contained only 34 ZFMK” material which has been obtained after 1971 from reptilian names documented by type material, 22 of which smaller university museums, first of all from the Zoolog- were name-bearing type specimens (onomatophores), and ical Museum of the University of Göttingen (1977). Sin- 12 further names were documented by paratypes only. -

Nghiên Cứu Đặc Điểm Hình Thái Và Sinh Thái Của Nhông Cát Leiolepis Guttata (Cuvier, 1829) Trong Điều Kiện Bán Hoang Dã Tại Huyện Bắc Bình, Tỉnh Bình Thuận
BỘ GIÁO DỤC VIỆN HÀN LÂM KHOA HỌC VÀ ĐÀO TẠO VÀ CÔNG NGHỆ VIỆT NAM HỌC VIỆN KHOA HỌC VÀ CÔNG NGHỆ ----------------------------- Nguyễn Thị Minh Phương NGHIÊN CỨU ĐẶC ĐIỂM HÌNH THÁI VÀ SINH THÁI CỦA NHÔNG CÁT LEIOLEPIS GUTTATA (CUVIER, 1829) TRONG ĐIỀU KIỆN BÁN HOANG DÃ TẠI HUYỆN BẮC BÌNH, TỈNH BÌNH THUẬN. LUẬN VĂN THẠC SĨ: SINH HỌC Thành phố Hồ Chí Minh, 2020 BỘ GIÁO DỤC VIỆN HÀN LÂM KHOA HỌC VÀ ĐÀO TẠO VÀ CÔNG NGHỆ VIỆT NAM HỌC VIỆN KHOA HỌC VÀ CÔNG NGHỆ ----------------------------- Nguyễn Thị Minh Phương NGHIÊN CỨU ĐẶC ĐIỂM HÌNH THÁI VÀ SINH THÁI CỦA NHÔNG CÁT LEIOLEPIS GUTTATA (CUVIER, 1829) TRONG ĐIỀU KIỆN BÁN HOANG DÃ TẠI HUYỆN BẮC BÌNH, TỈNH BÌNH THUẬN Chuyên ngành: Sinh học thực nghiệm Mã số: 8 42 01 14 LUẬN VĂN THẠC SĨ SINH HỌC NGƯỜI HƯỚNG DẪN KHOA HỌC: TS Trần Tình. Thành phố Hồ Chí Minh, 2020 LỜI CAM ĐOAN Tôi xin cam đoan đây là công trình nghiên cứu của bản thân tôi dưới sự hướng dẫn của TS. Trần Tình và tham khảo các tài liệu đã công bố có nguồn gốc rõ ràng. Các kết quả nêu trong luận văn hoàn toàn trung thực và chưa từng được bảo vệ trước bất kì hội đồng nào trước đây. Thành phố Hồ Chí Minh, tháng 7 năm 2020 Tác giả Nguyễn Thị Minh Phương LỜI CẢM ƠN Để hoàn thành được nội dung học tập và thực hiện luận văn này tôi xin tỏ lòng biết ơn đến Ban lãnh đạo Học viện Khoa học và Công nghệ, toàn thể quý thầy cô trong Học viện Khoa học và Công nghệ đã tận tình truyền giảng kiến thức, tư vấn kĩ thuật và hướng dẫn kĩ năng giúp tôi trau dồi nền tảng kiến thức cơ bản, thực hành kiến thức trong điều kiện thực tế. -

Dark Matter of Primate Genomes: Satellite DNA Repeats and Their Evolutionary Dynamics
cells Review Dark Matter of Primate Genomes: Satellite DNA Repeats and Their Evolutionary Dynamics Syed Farhan Ahmad 1,2, Worapong Singchat 1,2, Maryam Jehangir 1,3, Aorarat Suntronpong 1,2, Thitipong Panthum 1,2, Suchinda Malaivijitnond 4,5 and Kornsorn Srikulnath 1,2,4,6,7,* 1 Laboratory of Animal Cytogenetics and Comparative Genomics (ACCG), Department of Genetics, Faculty of Science, Kasetsart University, Bangkok 10900, Thailand; [email protected] (S.F.A.); [email protected] (W.S.); [email protected] (M.J.); [email protected] (A.S.); [email protected] (T.P.) 2 Special Research Unit for Wildlife Genomics (SRUWG), Department of Forest Biology, Faculty of Forestry, Kasetsart University, Bangkok 10900, Thailand 3 Department of Structural and Functional Biology, Institute of Bioscience at Botucatu, São Paulo State University (UNESP), Botucatu, São Paulo 18618-689, Brazil 4 National Primate Research Center of Thailand, Chulalongkorn University, Saraburi 18110, Thailand; [email protected] 5 Department of Biology, Faculty of Science, Chulalongkorn University, Bangkok 10330, Thailand 6 Center of Excellence on Agricultural Biotechnology (AG-BIO/PERDO-CHE), Bangkok 10900, Thailand 7 Omics Center for Agriculture, Bioresources, Food and Health, Kasetsart University (OmiKU), Bangkok 10900, Thailand * Correspondence: [email protected] Received: 27 October 2020; Accepted: 16 December 2020; Published: 18 December 2020 Abstract: A substantial portion of the primate genome is composed of non-coding regions, so-called “dark matter”, which includes an abundance of tandemly repeated sequences called satellite DNA. Collectively known as the satellitome, this genomic component offers exciting evolutionary insights into aspects of primate genome biology that raise new questions and challenge existing paradigms. -

Captive Wildlife Regulations, 2021, W-13.12 Reg 5
1 CAPTIVE WILDLIFE, 2021 W-13.12 REG 5 The Captive Wildlife Regulations, 2021 being Chapter W-13.12 Reg 5 (effective June 1, 2021). NOTE: This consolidation is not official. Amendments have been incorporated for convenience of reference and the original statutes and regulations should be consulted for all purposes of interpretation and application of the law. In order to preserve the integrity of the original statutes and regulations, errors that may have appeared are reproduced in this consolidation. 2 W-13.12 REG 5 CAPTIVE WILDLIFE, 2021 Table of Contents PART 1 PART 5 Preliminary Matters Zoo Licences and Travelling Zoo Licences 1 Title 38 Definition for Part 2 Definitions and interpretation 39 CAZA standards 3 Application 40 Requirements – zoo licence or travelling zoo licence PART 2 41 Breeding and release Designations, Prohibitions and Licences PART 6 4 Captive wildlife – designations Wildlife Rehabilitation Licences 5 Prohibition – holding unlisted species in captivity 42 Definitions for Part 6 Prohibition – holding restricted species in captivity 43 Standards for wildlife rehabilitation 7 Captive wildlife licences 44 No property acquired in wildlife held for 8 Licence not required rehabilitation 9 Application for captive wildlife licence 45 Requirements – wildlife rehabilitation licence 10 Renewal 46 Restrictions – wildlife not to be rehabilitated 11 Issuance or renewal of licence on terms and conditions 47 Wildlife rehabilitation practices 12 Licence or renewal term PART 7 Scientific Research Licences 13 Amendment, suspension, -
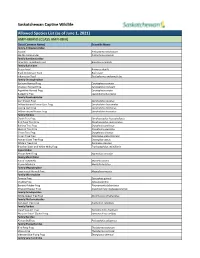
Captive Wildlife Allowed List
Saskatchewan Captive Wildlife Allowed Species List (as of June 1, 2021) AMPHIBIANS (CLASS AMPHIBIA) Class (Common Name) Scientific Name Family Ambystomatidae Axolotl Ambystoma mexicanum Marble Salamander Ambystoma opacum Family Bombinatoridae Oriental Fire-Bellied Toad Bombina orientalis Family Bufonidae Green Toad Anaxyrus debilis Black Indonesian Toad Bufo asper Indonesian Toad Duttaphrynus melanostictus Family Ceratophryidae Surinam Horned Frog Ceratophrys cornuta Chacoan Horned Frog Ceratophrys cranwelli Argentine Horned Frog Ceratophrys ornata Budgett’s Frog Lepidobatrachus laevis Family Dendrobatidae Dart Poison Frog Dendrobates auratus Yellow-banded Poison Dart Frog Dendrobates leucomelas Dyeing Dart Frog Dendrobates tinctorius Yellow-striped Poison Frog Dendrobates truncatus Family Hylidae Clown Tree Frog Dendropsophus leucophyllatus Bird Poop Tree Frog Dendropsophus marmoratus Barking Tree Frog Dryophytes gratiosus Squirrel Tree Frog Dryophytes squirellus Green Tree Frog Dryophytes cinereus Cuban Tree Frog Osteopilus septentrionalis Haitian Giant Tree Frog Osteopilus vastus White’s Tree Frog Ranoidea caerulea Brazilian Black and White Milky Frog Trachycephalus resinifictrix Hyperoliidae African Reed Frog Hyperolius concolor Family Mantellidae Baron’s Mantella Mantella baroni Brown Mantella Mantella betsileo Family Megophryidae Long-nosed Horned Frog Megophrys nasuta Family Microhylidae Tomato Frog Dyscophus guineti Chubby Frog Kaloula pulchra Banded Rubber Frog Phrynomantis bifasciatus Emerald Hopper Frog Scaphiophryne madagascariensis -

ZW Sex Chromosomes in Australian Dragon Lizards (Agamidae) Originated from a Combination of Duplication and Translocation in the Nucleolar Organising Region
G C A T T A C G G C A T genes Article ZW Sex Chromosomes in Australian Dragon Lizards (Agamidae) Originated from a Combination of Duplication and Translocation in the Nucleolar Organising Region 1, 1 1 1 Kazumi Matsubara y, Denis O’Meally , Stephen D. Sarre , Arthur Georges , Kornsorn Srikulnath 2 and Tariq Ezaz 1,* 1 Institute for Applied Ecology, Faculty of Science and Technology, University of Canberra, Canberra ACT 2617, Australia; [email protected] (K.M.); [email protected] (D.O.M.); [email protected] (S.D.S.); [email protected] (A.G.) 2 Department of Genetics, Faculty of Science, Kasetsart University, Bangkok 10900, Thailand; [email protected] * Correspondence: [email protected]; Tel.: +61-2-6201-2297 Current Address: Department of Biomedical Chemistry, Kwansei Gakuin University, Hyogo 669-1337, Japan. y Received: 23 September 2019; Accepted: 29 October 2019; Published: 30 October 2019 Abstract: Sex chromosomes in some reptiles share synteny with distantly related amniotes in regions orthologous to squamate chromosome 2. The latter finding suggests that chromosome 2 was formerly part of a larger ancestral (amniote) super-sex chromosome and raises questions about how sex chromosomes are formed and modified in reptiles. Australian dragon lizards (Agamidae) are emerging as an excellent model for studying these processes. In particular, they exhibit both genotypic (GSD) and temperature-dependent (TSD) sex determination, show evidence of transitions between the two modes and have evolved non-homologous ZW sex microchromosomes even within the same evolutionary lineage. They therefore represent an excellent group to probe further the idea of a shared ancestral super-sex chromosome and to investigate mechanisms for transition between different sex chromosome forms. -

Merilane Da Silva Calixto Mapeamento Cromossômico De DNA
Universidade Federal de Pernambuco Centro de Ciências Biológicas Programa de Pós-Graduacão em Genética Merilane da Silva Calixto Mapeamento cromossômico de DNA repetitivos em espécies de morcegos da família Phyllostomidae. Recife 2013 Merilane da Silva Calixto Mapeamento cromossômico de DNA repetitivos em espécies de morcegos da família Phyllostomidae. Tese apresentada ao Programa de Pós- Graduação em Genética da Universidade Federal de Pernambuco como parte dos requisitos exigidos para obtenção do título de Doutor em Genética. Orientadora: Dra. Maria José de Souza Lopes Coorientadora: Dra. Neide Santos Recife 2013 ii Catalogação na Fonte: Bibliotecário Bruno Márcio Gouveia, CRB-4/1788 Calixto, Merilane da Silva Mapeamento cromossômico de DNA repetitivos em espécies de morcegos da família Phyllostomidae / Merilane da Silva Calixto. – Recife: O Autor, 2013. 138 f.: il., fig., tab. Orientadora: Maria José de Souza Lopes Coorientadora: Neide Santos Tese (doutorado) – Universidade Federal de Pernambuco. Centro de Ciências Biológicas. Pós-graduação em Genética, 2013. Inclui bibliografia e anexos 1. Genética 2. DNA 3. Mapeamento cromossômico 4. Morcego I. Lopes, Maria José de Souza (orient.) II. Santos, Neide (coorient.) III. Título. 572.86 CDD (22.ed.) UFPE/CCB-2014-035 Merilane da Silva Calixto Mapeamento cromossômico de DNA repetitivos em espécies de morcegos da família Phyllostomidae Aprovado em 16/08/2013 Banca Examinadora: ____________________________________________ Dra. Maria José de Souza Lopes Universidade Federal de Pernambuco ____________________________________________ Dr. Martin Alejandro Montes Universidade Federal Rural de Pernambuco ____________________________________________ Dra. Katharine Raquel Pereira dos Santos Universidade Federal de Pernambuco ____________________________________________ Dra. Rita de Cássia de Moura Universidade de Pernambuco ____________________________________________ Dra. Tania Tassinari Rieger Universidade Federal de Pernambuco Recife 2013 iii Aos meus pais, às minhas irmãs e ao meu esposo Wellington, com amor dedico. -

Molecular Barcoding of Venomous Snakes and Species-Specific Multiplex PCR Assay to Identify Snake Groups for Which Antivenom Is Available in Thailand
Molecular barcoding of venomous snakes and species-specific multiplex PCR assay to identify snake groups for which antivenom is available in Thailand A. Supikamolseni1,2, N. Ngaoburanawit3, M. Sumontha4, L. Chanhome5, S. Suntrarachun6, S. Peyachoknagul2,7 and K. Srikulnath1,2,7 1Laboratory of Animal Cytogenetics and Comparative Genomics, Department of Genetics, Faculty of Science, Kasetsart University, Chatuchak, Bangkok, Thailand 2Department of Genetics, Faculty of Science, Kasetsart University, Chatuchak, Bangkok, Thailand 3Human Genetics Laboratory, Department of Pathology, Faculty of Medicine, Ramathibodi Hospital, Mahidol University, Bangkok, Thailand 4Ranong Marine Fisheries Station, Ranong, Thailand 5Snake Farm, Queen Saovabha Memorial Institute, The Thai Red Cross Society, Bangkok, Thailand 6Department of Research and Development, Queen Saovabha Memorial Institute, The Thai Red Cross Society, Bangkok, Thailand 7Center for Advanced Studies in Tropical Natural Resources, National Research University-Kasetsart University, Kasetsart University, Thailand Corresponding author: K. Srikulnath E-mail: [email protected] / [email protected] Genet. Mol. Res. 14 (4): 13981-13997 (2015) Received February 9, 2015 Accepted May 18, 2015 Published October 29, 2015 DOI http://dx.doi.org/10.4238/2015.October.29.18 ABSTRACT. DNA barcodes of mitochondrial COI and Cytb genes were constructed from 54 specimens of 16 species for species identification. Intra- and interspecific sequence divergence of the COI gene (10 times) was greater than that of the Cytb gene (4 times), which suggests that the Genetics and Molecular Research 14 (4): 13981-13997 (2015) ©FUNPEC-RP www.funpecrp.com.br A. Supikamolseni et al. 13982 former gene may be a better marker than the latter for species delimitation in snakes. -

ZOO REPORT PROFI the Content
No. 1 / march 2013 special supplement ZOO REPORT PROFI The Content The Speech Vladimir V. Spitsin Zooreport the magazine for friends of the Brno Zoo march 2013; No. 1/13, volume XV PAGE 3 The Largest Chameleon Michal Balcar Editor: Zoologická zahrada města Brna U Zoo 46, 635 00 Brno, Czech Republic tel.: +420 546 432 311 fax: +420 546 210 000 PAGE 4 e-mail: [email protected] Slavkovský Forest Protected Landscape Park RNDr. Pavel Řepa Publisher: Peleos, spol. s r.o. e-mail: [email protected] PAGE 5 Editor’s office address: Brno’s New Komodo Dragon Zoologická zahrada města Brna Michal Balcar Redakce Zooreport U Zoo 46, 635 00 Brno, Czech Republic tel.: +420 546 432 370 fax: +420 546 210 000 e-mail: [email protected] PAGES 6, 7 The City of Brno Has Assigned Hlídka to Zoo Editor manager: Eduard Stuchlík Bc. Eduard Stuchlík Specialist readers: PAGE 8 RNDr. Bohumil Král, CSc. Hot News Mgr. Lubomír Selinger (red) Emendation: Rosalind Miranda PAGE 9 Distribution: Kamchatka Brown Bears Love Honey and Snow 500 pcs in the English version Eduard Stuchlík 1,500 pcs in the Czech version Photos by: PAGE 10 Eduard Stuchlík Polar Bear Cora First page: is Taking Care of her Second Set of Twin Polar bears Eduard Stuchlík UNSALEABLE PAGE 11 2 The Speech EARAZA Organises Nine International Animal Protection Programmes The 19th annual conference of the Eurasian Regional Association of Zoos and Aquariums (EARAZA) will take place at the end of May 2013 at Brno Zoo. I would there- fore like to familiarize ZooReport readers with the association of which I have been the head since its foundation in 1994.