Molecular Barcoding of Venomous Snakes and Species-Specific Multiplex PCR Assay to Identify Snake Groups for Which Antivenom Is Available in Thailand
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

WHO Guidance on Management of Snakebites
GUIDELINES FOR THE MANAGEMENT OF SNAKEBITES 2nd Edition GUIDELINES FOR THE MANAGEMENT OF SNAKEBITES 2nd Edition 1. 2. 3. 4. ISBN 978-92-9022- © World Health Organization 2016 2nd Edition All rights reserved. Requests for publications, or for permission to reproduce or translate WHO publications, whether for sale or for noncommercial distribution, can be obtained from Publishing and Sales, World Health Organization, Regional Office for South-East Asia, Indraprastha Estate, Mahatma Gandhi Marg, New Delhi-110 002, India (fax: +91-11-23370197; e-mail: publications@ searo.who.int). The designations employed and the presentation of the material in this publication do not imply the expression of any opinion whatsoever on the part of the World Health Organization concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. Dotted lines on maps represent approximate border lines for which there may not yet be full agreement. The mention of specific companies or of certain manufacturers’ products does not imply that they are endorsed or recommended by the World Health Organization in preference to others of a similar nature that are not mentioned. Errors and omissions excepted, the names of proprietary products are distinguished by initial capital letters. All reasonable precautions have been taken by the World Health Organization to verify the information contained in this publication. However, the published material is being distributed without warranty of any kind, either expressed or implied. The responsibility for the interpretation and use of the material lies with the reader. In no event shall the World Health Organization be liable for damages arising from its use. -

Venom Protein of the Haematotoxic Snakes Cryptelytrops Albolabris
S HORT REPORT ScienceAsia 37 (2011): 377–381 doi: 10.2306/scienceasia1513-1874.2011.37.377 Venom protein of the haematotoxic snakes Cryptelytrops albolabris, Calloselasma rhodostoma, and Daboia russelii siamensis Orawan Khow, Pannipa Chulasugandha∗, Narumol Pakmanee Research and Development Department, Queen Saovabha Memorial Institute, Patumwan, Bangkok 10330 Thailand ∗Corresponding author, e-mail: pannipa [email protected] Received 1 Dec 2010 Accepted 6 Sep 2011 ABSTRACT: The protein concentration and protein pattern of crude venoms of three major haematotoxic snakes of Thailand, Cryptelytrops albolabris (green pit viper), Calloselasma rhodostoma (Malayan pit viper), and Daboia russelii siamensis (Russell’s viper), were studied. The protein concentrations of all lots of venoms studied were comparable. The chromatograms, from reversed phase high performance liquid chromatography, of C. albolabris venom and C. rhodostoma venom were similar but they were different from the chromatogram of D. r. siamensis venom. C. rhodostoma venom showed the highest number of protein spots on 2-dimensional gel electrophoresis (pH gradient 3–10), followed by C. albolabris venom and D. r. siamensis venom, respectively. The protein spots of C. rhodostoma venom were used as reference proteins in matching for similar proteins of haematotoxic snakes. C. albolabris venom showed more similar protein spots to C. rhodostoma venom than D. r. siamensis venom. The minimum coagulant dose could not be determined in D. r. siamensis venom. KEYWORDS: 2-dimensional gel electrophoresis, reverse phase high performance liquid chromatography, minimum coag- ulant dose INTRODUCTION inducing defibrination 5–7. The venom of D. r. sia- mensis directly affects factor X and factor V of the In Thailand there are 163 snake species, 48 of which haemostatic system 8,9 . -

Russell's Viper (Daboia Russelii) in Bangladesh: Its Boom and Threat To
J. Asiat. Soc. Bangladesh, Sci. 44(1): 15-22, June 2018 RUSSELL’S VIPER (DABOIA RUSSELII) IN BANGLADESH: ITS BOOM AND THREAT TO HUMAN LIFE MD. FARID AHSAN1* AND MD. ABU SAEED2 1Department of Zoology, University of Chittagong, Chittagong, Bangladesh 2 555, Kazipara, Mirpur, Dhaka-1216, Bangladesh Abstract The occurrence of Russell’s viper (Daboia russelii Shaw and Nodder 1797) in Bangladesh is century old information and its rarity was known to the wildlife biologists till 2013 but its recent booming is also causing a major threat to human life in the area. Recently it has been reported from nine districts (Dinajpur, Chapai Nawabganj, Rajshahi, Naogaon, Natore, Pabna, Rajbari, Chuadanga and Patuakhali) and old records revealed 11 districts (Nilphamari, Dinajpur, Rangpur, Chapai Nawabganj, Rajshahi, Bogra, Jessore, Satkhira, Khulna, Bagerhat and Chittagong). Thus altogether 17 out of 64 districts in Bangladesh, of which Chapai Nawabganj and Rajshahi are most affected and 20 people died due to Russell’s viper bite during 2013 to 2016. Its past and present distribution in Bangladesh and death toll of its bites have been discussed. Its booming causes have also been predicted and precautions have been recommended. Research on Russell’s viper is deemed necessary due to reemergence in deadly manner. Key words: Russell’s viper, Daboia russelii, Distribution, Boom, Panic, Death toll Introduction Two species of Russell’s viper are known to occur in this universe of which Daboia russelii (Shaw and Nodder 1797) is distributed in Pakistan, India, Nepal, Bhutan, Bangladesh and Sri Lanka (www.reptile.data-base.org); while Daboia siamensis (Smith 1917) occurs in China, Myanmar, Indonesia, Thailand, Taiwan and Cambodia (Wogan 2012). -

Venom Proteomics and Antivenom Neutralization for the Chinese
www.nature.com/scientificreports OPEN Venom proteomics and antivenom neutralization for the Chinese eastern Russell’s viper, Daboia Received: 27 September 2017 Accepted: 6 April 2018 siamensis from Guangxi and Taiwan Published: xx xx xxxx Kae Yi Tan1, Nget Hong Tan1 & Choo Hock Tan2 The eastern Russell’s viper (Daboia siamensis) causes primarily hemotoxic envenomation. Applying shotgun proteomic approach, the present study unveiled the protein complexity and geographical variation of eastern D. siamensis venoms originated from Guangxi and Taiwan. The snake venoms from the two geographical locales shared comparable expression of major proteins notwithstanding variability in their toxin proteoforms. More than 90% of total venom proteins belong to the toxin families of Kunitz-type serine protease inhibitor, phospholipase A2, C-type lectin/lectin-like protein, serine protease and metalloproteinase. Daboia siamensis Monovalent Antivenom produced in Taiwan (DsMAV-Taiwan) was immunoreactive toward the Guangxi D. siamensis venom, and efectively neutralized the venom lethality at a potency of 1.41 mg venom per ml antivenom. This was corroborated by the antivenom efective neutralization against the venom procoagulant (ED = 0.044 ± 0.002 µl, 2.03 ± 0.12 mg/ml) and hemorrhagic (ED50 = 0.871 ± 0.159 µl, 7.85 ± 3.70 mg/ ml) efects. The hetero-specifc Chinese pit viper antivenoms i.e. Deinagkistrodon acutus Monovalent Antivenom and Gloydius brevicaudus Monovalent Antivenom showed negligible immunoreactivity and poor neutralization against the Guangxi D. siamensis venom. The fndings suggest the need for improving treatment of D. siamensis envenomation in the region through the production and the use of appropriate antivenom. Daboia is a genus of the Viperinae subfamily (family: Viperidae), comprising a group of vipers commonly known as Russell’s viper native to the Old World1. -
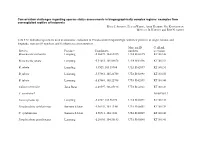
Conservation Challenges Regarding Species Status Assessments in Biogeographically Complex Regions: Examples from Overexploited Reptiles of Indonesia KYLE J
Conservation challenges regarding species status assessments in biogeographically complex regions: examples from overexploited reptiles of Indonesia KYLE J. SHANEY, ELIJAH WOSTL, AMIR HAMIDY, NIA KURNIAWAN MICHAEL B. HARVEY and ERIC N. SMITH TABLE S1 Individual specimens used in taxonomic evaluation of Pseudocalotes tympanistriga, with their province of origin, latitude and longitude, museum ID numbers, and GenBank accession numbers. Museum ID GenBank Species Province Coordinates numbers accession Bronchocela cristatella Lampung -5.36079, 104.63215 UTA R 62895 KT180148 Bronchocela jubata Lampung -5.54653, 105.04678 UTA R 62896 KT180152 B. jubata Lampung -5.5525, 105.18384 UTA R 62897 KT180151 B. jubata Lampung -5.57861, 105.22708 UTA R 62898 KT180150 B. jubata Lampung -5.57861, 105.22708 UTA R 62899 KT180146 Calotes versicolor Jawa Barat -6.49597, 106.85198 UTA R 62861 KT180149 C. versicolor* NC009683.1 Gonocephalus sp. Lampung -5.2787, 104.56198 UTA R 60571 KT180144 Pseudocalotes cybelidermus Sumatra Selatan -4.90149, 104.13401 UTA R 60551 KT180139 P. cybelidermus Sumatra Selatan -4.90711, 104.1348 UTA R 60549 KT180140 Pseudocalotes guttalineatus Lampung -5.28105, 104.56183 UTA R 60540 KT180141 P. guttalineatus Sumatra Selatan -4.90681, 104.13457 UTA R 60501 KT180142 Pseudocalotes rhammanotus Lampung -4.9394, 103.85292 MZB 10804 KT180147 Pseudocalotes species 4 Sumatra Barat -2.04294, 101.31129 MZB 13295 KT211019 Pseudocalotes tympanistriga Jawa Barat -6.74181, 107.0061 UTA R 60544 KT180143 P. tympanistriga Jawa Barat -6.74181, 107.0061 UTA R 60547 KT180145 Pogona vitticeps* AB166795.1 *Entry to GenBank by previous authors TABLE S2 Reptile species currently believed to occur Java and Sumatra, Indonesia, with IUCN Red List status, and certainty of occurrence. -

Evolutionary History of Spiny- Tailed Lizards (Agamidae: Uromastyx) From
Received: 6 July 2017 | Accepted: 4 November 2017 DOI: 10.1111/zsc.12266 ORIGINAL ARTICLE Evolutionary history of spiny- tailed lizards (Agamidae: Uromastyx) from the Saharo- Arabian region Karin Tamar1 | Margarita Metallinou1† | Thomas Wilms2 | Andreas Schmitz3 | Pierre-André Crochet4 | Philippe Geniez5 | Salvador Carranza1 1Institute of Evolutionary Biology (CSIC- Universitat Pompeu Fabra), Barcelona, The subfamily Uromastycinae within the Agamidae is comprised of 18 species: three Spain within the genus Saara and 15 within Uromastyx. Uromastyx is distributed in the 2Allwetterzoo Münster, Münster, Germany desert areas of North Africa and across the Arabian Peninsula towards Iran. The 3Department of Herpetology & systematics of this genus has been previously revised, although incomplete taxo- Ichthyology, Natural History Museum of nomic sampling or weakly supported topologies resulted in inconclusive relation- Geneva (MHNG), Geneva, Switzerland ships. Biogeographic assessments of Uromastycinae mostly agree on the direction of 4CNRS-UMR 5175, Centre d’Écologie Fonctionnelle et Évolutive (CEFE), dispersal from Asia to Africa, although the timeframe of the cladogenesis events has Montpellier, France never been fully explored. In this study, we analysed 129 Uromastyx specimens from 5 EPHE, CNRS, UM, SupAgro, IRD, across the entire distribution range of the genus. We included all but one of the rec- INRA, UMR 5175 Centre d’Écologie Fonctionnelle et Évolutive (CEFE), PSL ognized taxa of the genus and sequenced them for three mitochondrial and three Research University, Montpellier, France nuclear markers. This enabled us to obtain a comprehensive multilocus time- calibrated phylogeny of the genus, using the concatenated data and species trees. We Correspondence Karin Tamar, Institute of Evolutionary also applied coalescent- based species delimitation methods, phylogenetic network Biology (CSIC-Universitat Pompeu Fabra), analyses and model- testing approaches to biogeographic inferences. -

P. 1 AC27 Inf. 7 (English Only / Únicamente En Inglés / Seulement
AC27 Inf. 7 (English only / únicamente en inglés / seulement en anglais) CONVENTION ON INTERNATIONAL TRADE IN ENDANGERED SPECIES OF WILD FAUNA AND FLORA ____________ Twenty-seventh meeting of the Animals Committee Veracruz (Mexico), 28 April – 3 May 2014 Species trade and conservation IUCN RED LIST ASSESSMENTS OF ASIAN SNAKE SPECIES [DECISION 16.104] 1. The attached information document has been submitted by IUCN (International Union for Conservation of * Nature) . It related to agenda item 19. * The geographical designations employed in this document do not imply the expression of any opinion whatsoever on the part of the CITES Secretariat or the United Nations Environment Programme concerning the legal status of any country, territory, or area, or concerning the delimitation of its frontiers or boundaries. The responsibility for the contents of the document rests exclusively with its author. AC27 Inf. 7 – p. 1 Global Species Programme Tel. +44 (0) 1223 277 966 219c Huntingdon Road Fax +44 (0) 1223 277 845 Cambridge CB3 ODL www.iucn.org United Kingdom IUCN Red List assessments of Asian snake species [Decision 16.104] 1. Introduction 2 2. Summary of published IUCN Red List assessments 3 a. Threats 3 b. Use and Trade 5 c. Overlap between international trade and intentional use being a threat 7 3. Further details on species for which international trade is a potential concern 8 a. Species accounts of threatened and Near Threatened species 8 i. Euprepiophis perlacea – Sichuan Rat Snake 9 ii. Orthriophis moellendorfi – Moellendorff's Trinket Snake 9 iii. Bungarus slowinskii – Red River Krait 10 iv. Laticauda semifasciata – Chinese Sea Snake 10 v. -

An Epidemiological Study of Venomous Snake Bites: a Hospital Based Analysis
ORIGINAL RESEARCH ARTICLE pISSN 0976 3325│eISSN 2229 6816 Open Access Article www.njcmindia.org An Epidemiological Study of Venomous Snake Bites: A Hospital Based Analysis Dipak H. Vora1, Jyoti H. Vora2 Financial Support: None declared ABSTRACT Conflict of Interest: None declared Copy Right: The Journal retains the Background & Objectives: This study was carried out to describe copyrights of this article. However, re- epidemiology of snake bite cases which were seen in a tertiary care production is permissible with due ac- hospital of Ahmedabad region so that the data provided will help knowledgement of the source. in estimating the envenoming in this region of India. How to cite this article: Methods: Total 50 cases of venomous snake bites were studied ret- Vora DH, Vora JH. An Epidemiological rospectively. These patients were admitted in the Medicine De- Study of Venomous Snake Bites: A partment of V.S. Hospital, Ahmedabad during the period from Hospital Based Analysis. Natl J Com- April 2008 to October 2009. munity Med 2019;10(8):474-478 Results: Maximum number of cases (66 %) was belonging to the Author’s Affiliation: age group 15-34 years. Male are having twice the incidence com- 1 Assistant Professor, Department of pare to the female (M: F ratio 2.12:1). Maximum cases were from Forensic Medicine, B. J. Medical Col- rural areas i.e. 72 %. In 66 % cases, snake bites occurred during lege; 2Associate Professor, Department of Medicine, SMT NHL Municipal night time. Most of the cases i.e. 82 % occurred during rainy sea- Medical College, Ahmedabad son. Elapid snake bites leading to neurotoxicity is common fol- lowed by viperidae induced vasculotoxicity and acute renal fail- Correspondence ure. -

Zootaxa 2433: 47–61 (2010) ISSN 1175-5326 (Print Edition) Article ZOOTAXA Copyright © 2010 · Magnolia Press ISSN 1175-5334 (Online Edition)
Zootaxa 2433: 47–61 (2010) ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Article ZOOTAXA Copyright © 2010 · Magnolia Press ISSN 1175-5334 (online edition) Who’s your mommy? Identifying maternal ancestors of asexual species of Leiolepis Cuvier, 1829 and the description of a new endemic species of asexual Leiolepis Cuvier, 1829 from Southern Vietnam JESSE L. GRISMER1 & L. LEE GRISMER2 1Department of Biology Villanova University, 800 Lancaster Ave, Villanova, Pennsylvania, 19085 USA E-mail: [email protected] 2Department of Biology La Sierra University, Riverside, 4500 Riverwalk Parkway, California, 92515 USA Abstract A new asexual species of Leiolepis is described from Binh Chau – Phuoc Buu Nature Reserve, Xuyen Moc district Ba Ria-Vung Tau Province, Vietnam to where it is believed to be endemic. Leiolepis ngovantrii sp. nov. differs from all sexual species of Leiolepis by lacking males and from all asexual species by having nine rows of enlarged keeled scales across the forearm and 37–40 subdigital lamellae beneath the fourth toe. Phylogenetic inference based on 700 base pairs of the mitochondrial ND2 region, placed L. ngovantrii sp. nov. among the currently described asexual species and was used to assess the maternal ancestors of the remaining asexual species. Both maximum parsimony and maximum likelihood analyses recovered L. guttata as the maternal ancestors of L. guentherpetersi, L. boehmei, and L. ngovantrii sp. nov., and L. boehmei as the maternal ancestor to L. triploida. Key words: asexual, Leiolepis, maternal, ngovantrii, new species, taxonomy, Vietnam Introduction The discovery of asexual, vertebrate lineages (Darevsky, 1967) has opened the door to a new chapter of evolutionary biology that is still not completely understood (Schön et al., 2009). -

Revised Distribution and a First Record of Leiolepis Peguensis Peters, 1971 (Squamata: Leiolepidae) from Thailand
Herpetology Notes, volume 14: 893-897 (2021) (published online on 10 June 2021) Revised distribution and a first record of Leiolepis peguensis Peters, 1971 (Squamata: Leiolepidae) from Thailand Pattarapon Promnun1,*, Jenjit Khudamrongsawat1, Jesse L. Grismer2, Nontivich Tandavanitj3, Chalita Kongrit1, and Preecha Tajakan4 The Butterfly lizard, Leiolepis peguensis Peters, 1971, and collected tissue samples (tail tips). Three specimens is terrestrial, diurnal, and omnivorous. It possesses unique were deposited in the Thailand Natural History Museum, external body patterns (Peters, 1971) that distinguish it Pathum Thani, Thailand (voucher no. THNHM28601– from all other species of Leiolepis and has equally been 28603), while other specimens were released after shown to be genetically distinct (Grismer and Grismer, examination. They were identified to species level 2010; Grismer et al., 2014). It was previously reported based on meristic and molecular approaches. in Pegu-Yoma on the eastern side of Irrawaddy River, Meristic characteristics of our specimens were from Arakan-Yoma and Chatthin Wildlife Sanctuary on compared to those of the following Leiolepis species: L. western side of the Irrawaddy River (Shan State), and some localities in central and west of Myanmar (Peters, 1971; Zug et al., 1998; Grismer et al., 2014). Although L. peguensis is common in Myanmar, it has never been reported east of the Tenasserim mountain range which geographically separates Myanmar from the rest of Southeast Asia. Here we present a new record of L. peguensis from east of the Tenasserim range in Mae Hong Son Province, northern Thailand (Fig. 1). We encountered five adult L. peguensis (Fig. 2) in Pang Mu Sub-district (19.3572°N, 97.9809°E) and Pha Bong Sub-district (19.2394°N, 97.9978°E), Namtok Mae Surin National Park, during two dry seasons (March–May) in 2018–2019. -

Genetic Relationship of Three Butterfly Lizard Species (Leiolepis Reevesii Rubritaeniata, Leiolepis Belliana Belliana, Leiolepis
Kasetsart J. (Nat. Sci.) 44 : 424 - 435 (2010) Genetic Relationship of Three Butterfly Lizard Species (Leiolepis reevesii rubritaeniata, Leiolepis belliana belliana, Leiolepis boehmei, Agamidae, Squamata) Inferred from Nuclear Gene Sequence Analyses Kornsorn Srikulnath1, 2, Kazumi Matsubara3, Yoshinobu Uno2, Amara Thongpan1, Saowanee Suputtitada1, Chizuko Nishida2, 3, Yoichi Matsuda2, 3, 4 and Somsak Apisitwanich1* ABSTRACT The genetic relationship was investigated of three butterfly lizard species (Leiolepis reevesii rubritaeniata, L. belliana belliana and L. boehmei) selectively inhabiting Thailand. The findings were based on RAG1 and C-mos gene analyses. The DNA sequences were also compared with the other squamate reptiles. The analysis strongly supported that L. reevesii rubritaeniata was related more closely to L. belliana belliana than to L. boehmei. The phylogenetic position of Leiolepis spp., however, was contentious with regard to its relationship among the Leiolepidinae, Agaminae and Chamaeleonidae, which suggested that their phylogeny remains uncertain. Keywords: butterfly lizard, Leiolepidinae, phylogeny, RAG1, C-mos INTRODUCTION inhabit Southeast Asia. They show a great variety of karyotypes and sexual systems. In Thailand, The Squamata is the most diverse there are three species, which barely can be reptilian order that has been classified traditionally discriminated from other congeneric species by into three suborders: Serpentes (snakes), their typical scale and skin coloration (Peters, Amphisbaenia (worm lizards) and Lacertilia 1971). Bisexualism has been described in Leiolepis (lizards). The extant lizards can be further belliana belliana (2n=2x=36), which is widely categorized into five infraorders (the Iguania, found throughout the country, L. belliana ocellata Gekkota, Scincomorpha, Diploglossa, Dibamia, (2n=2x=34) found in upper northern, and L. -

The Oligocene and Miocene Fossil Lizards (Reptilia, Squamata) of Central Mongolia
geodiversitas 2019 ● 41 ● 24 e of lif pal A eo – - e h g e r a p R e t e o d l o u g a l i s C - t – n a M e J e l m a i r o DIRECTEUR DE LA PUBLICATION : Bruno David, Président du Muséum national d’Histoire naturelle RÉDACTEUR EN CHEF / EDITOR-IN-CHIEF : Didier Merle ASSISTANT DE RÉDACTION / ASSISTANT EDITOR : Emmanuel Côtez ([email protected]) MISE EN PAGE / PAGE LAYOUT : Emmanuel Côtez COMITÉ SCIENTIFIQUE / SCIENTIFIC BOARD : Christine Argot (MNHN, Paris) Beatrix Azanza (Museo Nacional de Ciencias Naturales, Madrid) Raymond L. Bernor (Howard University, Washington DC) Alain Blieck (chercheur CNRS retraité, Haubourdin) Henning Blom (Uppsala University) Jean Broutin (UPMC, Paris) Gaël Clément (MNHN, Paris) Ted Daeschler (Academy of Natural Sciences, Philadelphie) Bruno David (MNHN, Paris) Gregory D. Edgecombe (The Natural History Museum, Londres) Ursula Göhlich (Natural History Museum Vienna) Jin Meng (American Museum of Natural History, New York) Brigitte Meyer-Berthaud (CIRAD, Montpellier) Zhu Min (Chinese Academy of Sciences, Pékin) Isabelle Rouget (UPMC, Paris) Sevket Sen (MNHN, Paris) Stanislav Štamberg (Museum of Eastern Bohemia, Hradec Králové) Paul Taylor (The Natural History Museum, Londres) COUVERTURE / COVER : Made from the Figures of the article. Geodiversitas est indexé dans / Geodiversitas is indexed in: – Science Citation Index Expanded (SciSearch®) – ISI Alerting Services® – Current Contents® / Physical, Chemical, and Earth Sciences® – Scopus® Geodiversitas est distribué en version électronique par / Geodiversitas is distributed