Functional MRI Reveals Locomotion-Control Neural Circuits in Human Brainstem
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

NS201C Anatomy 1: Sensory and Motor Systems
NS201C Anatomy 1: Sensory and Motor Systems 25th January 2017 Peter Ohara Department of Anatomy [email protected] The Subdivisions and Components of the Central Nervous System Axes and Anatomical Planes of Sections of the Human and Rat Brain Development of the neural tube 1 Dorsal and ventral cell groups Dermatomes and myotomes Neural crest derivatives: 1 Neural crest derivatives: 2 Development of the neural tube 2 Timing of development of the neural tube and its derivatives Timing of development of the neural tube and its derivatives Gestational Crown-rump Structure(s) age (Weeks) length (mm) 3 3 cerebral vesicles 4 4 Optic cup, otic placode (future internal ear) 5 6 cerebral vesicles, cranial nerve nuclei 6 12 Cranial and cervical flexures, rhombic lips (future cerebellum) 7 17 Thalamus, hypothalamus, internal capsule, basal ganglia Hippocampus, fornix, olfactory bulb, longitudinal fissure that 8 30 separates the hemispheres 10 53 First callosal fibers cross the midline, early cerebellum 12 80 Major expansion of the cerebral cortex 16 134 Olfactory connections established 20 185 Gyral and sulcul patterns of the cerebral cortex established Clinical case A 68 year old woman with hypertension and diabetes develops abrupt onset numbness and tingling on the right half of the face and head and the entire right hemitrunk, right arm and right leg. She does not experience any weakness or incoordination. Physical Examination: Vitals: T 37.0° C; BP 168/87; P 86; RR 16 Cardiovascular, pulmonary, and abdominal exam are within normal limits. Neurological Examination: Mental Status: Alert and oriented x 3, 3/3 recall in 3 minutes, language fluent. -

Magnetic Resonance Imaging Techniques for Visualization of the Subthalamic Nucleus
J Neurosurg 115:971–984, 2011 Magnetic resonance imaging techniques for visualization of the subthalamic nucleus A review ELLEN J. L. BRUNENbeRG, M.SC.,1 BRAM PLATEL, PH.D.,2 PAUL A. M. HOFMAN, PH.D., M.D.,3 BART M. TER HAAR ROmeNY, PH.D.,1,4 AND VeeRLE VIsseR-VANdeWALLE, PH.D., M.D.5,6 1Department of Biomedical Engineering, Eindhoven University of Technology, Eindhoven; Departments of 3Radiology, 4Biomedical Engineering, and 5Neurosurgery, and 6Maastricht Institute for Neuromodulative Development, Maastricht University Medical Center, Maastricht, The Netherlands; and 2Fraunhofer MEVIS, Bremen, Germany The authors reviewed 70 publications on MR imaging–based targeting techniques for identifying the subtha- lamic nucleus (STN) for deep brain stimulation in patients with Parkinson disease. Of these 70 publications, 33 presented quantitatively validated results. There is still no consensus on which targeting technique to use for surgery planning; methods vary greatly between centers. Some groups apply indirect methods involving anatomical landmarks, or atlases incorporating ana- tomical or functional data. Others perform direct visualization on MR imaging, using T2-weighted spin echo or inver- sion recovery protocols. The combined studies do not offer a straightforward conclusion on the best targeting protocol. Indirect methods are not patient specific, leading to varying results between cases. On the other hand, direct targeting on MR imaging suffers from lack of contrast within the subthalamic region, resulting in a poor delineation of the STN. These defi- ciencies result in a need for intraoperative adaptation of the original target based on test stimulation with or without microelectrode recording. It is expected that future advances in MR imaging technology will lead to improvements in direct targeting. -
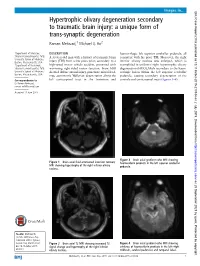
Hypertrophic Olivary Degeneration Secondary to Traumatic Brain Injury: a Unique Form of Trans-Synaptic Degeneration Raman Mehrzad,1 Michael G Ho2
… Images in BMJ Case Reports: first published as 10.1136/bcr-2015-210334 on 2 July 2015. Downloaded from Hypertrophic olivary degeneration secondary to traumatic brain injury: a unique form of trans-synaptic degeneration Raman Mehrzad,1 Michael G Ho2 1Department of Medicine, DESCRIPTION haemorrhagic left superior cerebellar peduncle, all Steward Carney Hospital, Tufts A 33-year-old man with a history of traumatic brain consistent with his prior TBI. Moreover, the right University School of Medicine, Boston, Massachusetts, USA injury (TBI) from a few years prior, secondary to a inferior olivary nucleus was enlarged, which is 2Department of Neurology, high-speed motor vehicle accident, presented with exemplified in unilateral right hypertrophic olivary Steward Carney Hospital, Tufts worsening right-sided motor function. Brain MRI degeneration (HOD), likely secondary to the haem- University School of Medicine, showed diffuse axonal injury, punctuate microbleed- orrhagic lesion within the left superior cerebellar Boston, Massachusetts, USA ings, asymmetric Wallerian degeneration along the peduncle, causing secondary degeneration of the fi – Correspondence to left corticospinal tract in the brainstem and contralateral corticospinal tracts ( gures 1 6). Dr Raman Mehrzad, [email protected] Accepted 11 June 2015 http://casereports.bmj.com/ fl Figure 3 Brain axial gradient echo MRI showing Figure 1 Brain axial uid-attenuated inversion recovery haemosiderin products in the left superior cerebellar MRI showing hypertrophy of the right inferior olivary peduncle. nucleus. on 25 September 2021 by guest. Protected copyright. To cite: Mehrzad R, Ho MG. BMJ Case Rep Published online: [please include Day Month Year] Figure 2 Brain axial T2 MRI showing increased T2 Figure 4 Brain axial gradient echo MRI showing doi:10.1136/bcr-2015- signal change and hypertrophy of the right inferior evidence of haemosiderin products in the left>right 210334 olivary nucleus. -

Closed-Loop Neuromodulation Restores Network Connectivity And
RESEARCH ARTICLE Closed-loop neuromodulation restores network connectivity and motor control after spinal cord injury Patrick D Ganzer1,2*, Michael J Darrow1, Eric C Meyers1,2, Bleyda R Solorzano2, Andrea D Ruiz2, Nicole M Robertson2, Katherine S Adcock3, Justin T James2, Han S Jeong2, April M Becker4, Mark P Goldberg4, David T Pruitt1,2, Seth A Hays1,2,3, Michael P Kilgard1,2,3, Robert L Rennaker II1,2,3* 1Erik Jonsson School of Engineering and Computer Science, The University of Texas at Dallas, Richardson, United States; 2Texas Biomedical Device Center, Richardson, United States; 3School of Behavioral Brain Sciences, The University of Texas at Dallas, Richardson, United States; 4Department of Neurology and Neurotherapeutics, University of Texas Southwestern Medical Center, Dallas, United States Abstract Recovery from serious neurological injury requires substantial rewiring of neural circuits. Precisely-timed electrical stimulation could be used to restore corrective feedback mechanisms and promote adaptive plasticity after neurological insult, such as spinal cord injury (SCI) or stroke. This study provides the first evidence that closed-loop vagus nerve stimulation (CLV) based on the synaptic eligibility trace leads to dramatic recovery from the most common forms of SCI. The addition of CLV to rehabilitation promoted substantially more recovery of forelimb function compared to rehabilitation alone following chronic unilateral or bilateral cervical SCI in a rat model. Triggering stimulation on the most successful movements is critical to maximize recovery. CLV enhances recovery by strengthening synaptic connectivity from remaining motor *For correspondence: networks to the grasping muscles in the forelimb. The benefits of CLV persist long after the end of [email protected] stimulation because connectivity in critical neural circuits has been restored. -

Metabolic Activity of Red Nucleus and Its Correlation with Cerebral Cortex and Cerebellum: a Study Using a High-Resolution Semiconductor PET System
Metabolic Activity of Red Nucleus and Its Correlation with Cerebral Cortex and Cerebellum: A Study Using a High-Resolution Semiconductor PET System Kenji Hirata1,2, Naoya Hattori3, Wataru Takeuchi4, Tohru Shiga1, Yuichi Morimoto4, Kikuo Umegaki5, Kentaro Kobayashi1, Osamu Manabe1, Shozo Okamoto1, and Nagara Tamaki1 1Department of Nuclear Medicine, Hokkaido University Graduate School of Medicine, Sapporo, Japan; 2Department of Molecular and Medical Pharmacology, David Geffen School of Medicine at UCLA, Los Angeles, California; 3Department of Molecular Imaging, Hokkaido University Graduate School of Medicine, Sapporo, Japan; 4Research and Development Group, Hitachi Ltd., Tokyo, Japan; and 5Division of Quantum Science and Engineering, Faculty of Engineering, Hokkaido University, Sapporo, Japan (1). The rubrospinal tract mainly controls limb musculature, whereas The red nucleus (RN) is a pair of small gray matter structures located the pyramidal tract can act on the whole musculature (1). A recent in the midbrain and involved in muscle movement and cognitive anatomic study indicated that a significant amount of rubrospinal tract functions. This retrospective study aimed to investigate the metabo- is present in the human brain (2). Because neuronal activity of the RN lism of human RN and its correlation to other brain regions. Methods: in Parkinson disease is known to be increased during passive and We developed a high-resolution semiconductor PET system to image voluntary movements (3), the RN may play a role in the coordination small brain structures. Twenty patients without neurologic disorders of muscular movement. Cognitive symptoms, such as intellectual underwent whole-brain scanning after injection of 400 MBq of fatigability, decreased verbal fluency, and discrete memory impair- 18F-FDG. -

Compensatory Sprouting and Impulse Rerouting After Unilateral Pyramidal Tract Lesion in Neonatal Rats
The Journal of Neuroscience, September 1, 2000, 20(17):6561–6569 Compensatory Sprouting and Impulse Rerouting after Unilateral Pyramidal Tract Lesion in Neonatal Rats Werner J. Z’Graggen, Karim Fouad, Olivier Raineteau, Gerlinde A. S. Metz, Martin E. Schwab, and Gwendolyn L. Kartje Brain Research Institute, University of Zurich and Swiss Federal Institute of Technology Zurich, CH-8057 Zurich, Switzerland After lesions of the developing mammalian CNS, structural plas- tigate possible new functional connections from the motor cortex ticity and functional recovery are much more pronounced than in of the pyramidotomy side to the periphery. In rats lesioned as the mature CNS. We investigated the anatomical reorganization adults, stimulation of the motor cortex ipsilateral to the pyra- of the corticofugal projections rostral to a unilateral lesion of the midotomy never elicited EMG activity. In contrast, in P2 lesioned corticospinal tract at the level of the medullary pyramid (pyra- rats bilateral forelimb EMGs were found. EMG latencies were midotomy) and the contribution of this reorganization and other comparable for the ipsilateral and contralateral responses but descending systems to functional recovery. were significantly longer than in unlesioned animals. Transient Two-day-old (P2) and adult rats underwent a unilateral pyra- inactivation of both red nuclei with the GABA receptor agonist midotomy. Three months later the corticofugal projections to the muscimol led to a complete loss of these bilateral movements. red nucleus and the pons were analyzed; a relatively large num- Movements and EMGs reappeared after wash-out of the drug. ber of corticorubral and corticopontine fibers from the lesioned These results suggest an important role of the red nucleus in the side had crossed the midline and established an additional con- tralateral innervation of the red nucleus and the pons. -

Identification of the Rubro-Olivary Tract in the Human Brain: a Diffusion Tensor Tractography Study
Original Article J. Phys. Ther. Sci. 22: 7–10, 2010 Identification of the Rubro-Olivary Tract in the Human Brain: a Diffusion Tensor Tractography Study SUNG HO JANG, MD1), JI HEON HONG, PT, MS1), YONG HYUN KWON, PT, PhD2) 1)Department of Physical Medicine and Rehabilitation, College of Medicine, Yeungnam University 2)Department of Physical Therapy, Yeungnam College of Science & Technology: 1737, Daemyung 7-Dong, Namgu, Daegu, 705-703, Republic of Korea. TEL +82 53-650-9702, FAX: +82 53-629-5048 Abstract. [Purpose] Little is known about the rubro-olivary tract (ROT) in the human brain. We attempted to identify the ROT using diffusion tensor tractography (DTT). [Subjects and Methods] We obtained ROT data from 11 healthy subjects with no history of neurological disorder. For tracking of the ROT, a seed region of interest (ROI) was selected in the red nucleus, and a target ROI was found in the inferior olive of each subject. [Results] The ROT, originated in the red nucleus, and passed laterally to the decussation of the superior cerebellar peduncle in the lower midbrain. In the pons, it descended through the area adjacent to the medial lemnicus in the posterior direction. Within the medulla, the ROT ended in the inferior olive, which was located lateral to the medial lemnicus and posterior to the pyramid. [Conclusion] We identified the ROT in the human brain using DTT. These results will be informative for research into the ROT in the human brain. Key words: Diffusion tensor image, Red nucleus, Inferior olive (This article was submitted Jul. 24, 2009, and was accepted Sep. -

Midbrain Tegmental Lesions Affecting Or Sparing the Pupillary Fibres
J7ournal ofNeurology, Neurosurgery, and Psychiatry 1996;61:401-402 401 J Neurol Neurosurg Psychiatry: first published as 10.1136/jnnp.61.4.401 on 1 October 1996. Downloaded from SHORT REPORT Midbrain tegmental lesions affecting or sparing the pupillary fibres Naokatsu Saeki, Naohisa Murai, Kenro Sunami Abstract lesion in the upper midbrain and close to the Two patients with oculomotor palsy third ventricle (fig 1). caused by midbrain infarction are Three months later the oculomotor palsy reported. In the first, pupillary reaction improved. The patient returned to his previ- was affected and in the second this reac- ous work after a further three months. tion was spared. Because the lesions in the anterior part of the tegmentum were CASE 2 in the upper midbrain in the first patient A 68 year old woman with hypertension for and in the lower midbrain in the second, eight years suddenly developed vertigo and it is suggested that the pupillary compo- nents of the oculomotor nerve are located in the upper midbrain. (7 Neurol Neurosurg Psychiatry 1996;61:401-402) Keywords: midbrain; oculomotor nerve; pupil sparing We report the details of two patients with a small midbrain infarction, the first with impairment of pupillary reaction to light and the second in which this reaction was pre- served. The aim of this study was to elucidate the topography of oculomotor pupillary fibres in the midbrain tegmentum based on findings using MRI. http://jnnp.bmj.com/ Case studies CASE 1 A 67 year old man with a 10 year history of hypertension presented with difficulty in open- ing his left eye on waking up in the morning. -

Axotomy-Induced Alterations in the Red Nucleus Revealed by Monoclonal Antibody, Py, Following a Low Thoracic Spinal Cord Lesion in the Adult Rat
Spinal Cord (1997) 35, 474 ± 481 1997 International Medical Society of Paraplegia All rights reserved 1362 ± 4393/97 $12.00 Axotomy-induced alterations in the red nucleus revealed by monoclonal antibody, Py, following a low thoracic spinal cord lesion in the adult rat GA Brook1, W Nacimiento1, A-S Taheri1, PL Woodhams2 and J Noth1 1Department of Neurology, Aachen University Medical School, Pauwelsstraûe 30, 52057-Aachen, Germany; 2Norman and Sadie Lee Research Centre, Division of Neurobiology, National Institute for Medical Research, Mill Hill, London NW7 1AA, UK The monoclonal antibody Py was previously developed as a tool for the identi®cation of sub- populations of hippocampal neurons. Here, the dierential distribution of Py immunore- activity in the mid-brain is described showing that Py also serves as a useful marker for other populations of neurons. Medium to strong immunoreactivity was observed in the cell body and dendrites of neurons of the oculomotor nucleus, superior colliculus and substantia gelatinosa reticulata. However, particularly intense Py-immunoreactivity was identi®ed in the magnocellular neurons in the caudal pole of the red nucleus. Unilateral transection of the rubrospinal tract at Th9-10 induced a marked reduction of Py immunoreactivity in the ventrolateral territory of the caudal pole of the axotomised red nucleus. A small but statistically signi®cant reduction of Py-immunoreactivity was ®rst seen at 7 days after surgery and a maximal loss of immunoreactivity (reduced to 66% of control levels) was observed by 21 days after surgery. Immunoreactivity in the axotomised red nucleus was reduced for the duration of the experiment but at the longer survival times studied (3 and 6 months) a small degree of recovery of staining was observed in small-medium diameter atrophic neurons. -

The Pedunculopontine Nucleus Area: Critical Evaluation of Interspecies Differences Relevant
doi:10.1093/brain/awq322 Brain 2011: 134; 11–23 | 11 BRAIN A JOURNAL OF NEUROLOGY REVIEW ARTICLE The pedunculopontine nucleus area: critical evaluation of interspecies differences relevant for its use as a target for deep brain stimulation Downloaded from https://academic.oup.com/brain/article/134/1/11/293572 by guest on 23 September 2021 Mesbah Alam, Kerstin Schwabe and Joachim K. Krauss Department of Neurosurgery, Medical University of Hannover, Hannover 30625, Germany Correspondence to: Prof. Dr. med. Joachim K. Krauss, Department of Neurosurgery, Medical University of Hannover, Carl-Neuberg-Str. 1, 30625 Hannover, Germany E-mail: [email protected] Recently, the pedunculopontine nucleus has been highlighted as a target for deep brain stimulation for the treatment of freezing of postural instability and gait disorders in Parkinson’s disease and progressive supranuclear palsy. There is great controversy, however, as to the exact location of the optimal site for stimulation. In this review, we give an overview of anatomy and connectivity of the pedunculopontine nucleus area in rats, cats, non-human primates and humans. Additionally, we report on the behavioural changes after chemical or electrical manipulation of the pedunculopontine nucleus. We discuss the relation to adjacent regions of the pedunculopontine nucleus, such as the cuneiform nucleus and the subcuneiform nucleus, which together with the pedunculopontine nucleus are the main areas of the mesencephalic locomotor region and play a major role in the initiation of gait. This information is discussed with respect to the experimental designs used for research purposes directed to a better understanding of the circuitry pathway of the pedunculopontine nucleus in association with basal ganglia pathology, and with respect to deep brain stimulation of the pedunculopontine nucleus area in humans. -

Rubrospinal Tract
LECTURE IV: Physiology of Motor Tracts EDITING FILE GSLIDES IMPORTANT MALE SLIDES EXTRA FEMALE SLIDES LECTURER’S NOTES 1 PHYSIOLOGY OF MOTOR TRACTS Lecture Four In order to initiate any type of voluntary movement there will be 2 levels of neuron that your body will use and they are: Upper Motor Neurons (UMN) Lower Motor Neurons (LMN) These are the motor These are the motor neurons whose cell bodies neurons of the spinal lie in the motor cortex, or cord (AHCs) and brain brainstem, and they stem motor nuclei of the activate the lower motor cranial nerves that neuron innervates skeletal muscle directly. Figure 4-1 The descending motor system (pyramidal,Extrapyramidal )has a number of important sets these are named according to the origin of their cell bodies and their final destination; Originates from the cerebral ● The rest of the descending motor pathways 1 cortex and descends to the pyramidal do not travel through the medullary pyramids spinal cord (the corticospinal extra and are therefore collectively gathered under tract) passes through the the heading:“the extrapyramidal tracts” pyramids of the medulla and ● Responsible for subconscious gross therefore has been called the “the pyramidal movements(swinging of arms during walking) pyramidal tract ” DESCENDING MOTOR SYSTEM PYRAMIDAL EXTRAPYRAMIDAL Corticospinal Corticobulbar Rubrospinal Vestibulospinal Tectospinal tracts tracts tracts tracts tracts Reticulospinal Olivospinal tract Tract FOOTNOTES 1. They are collections of white matter in the medulla that appear triangular due to crossing of motor tracts. Therefore they are termed “medullary pyramids”. 2 PHYSIOLOGY OF MOTOR TRACTS Lecture Four MOTOR AREAS Occupies the Precentral Area of representation Gyrus & contains large, is proportional with the giant highly excitable complexity of function Betz cells. -

A Patient with Bilateral Tremors Secondary to a Unilateral Brainstem Lesion: the Utility of Mollaret’S Triangle
Case Report Annals of Clinical Case Reports Published: 05 Jul, 2016 A Patient with Bilateral Tremors Secondary to a Unilateral Brainstem Lesion: The Utility of Mollaret’s Triangle Tran AT1*, Deep A2, Moguel-Cobos G2 and Lieberman A2 1Department of Neurology, U.S. Department of Veterans Affairs, USA 2Department of Neurology, Barrow Neurological Institute, USA Abstract A unilateral tremor developed in a patient’s left arm after a right midbrain hemorrhage. Thirteen years later, a re-bleed into that same area caused an additional right arm tremor. He now had bilateral arm tremors from a unilateral midbrain hemorrhage. The tremor was refractory to medications (propranolol, primidone, clonazepam, and levodopa). MRI brain showed bilateral hypertrophic olivary degeneration (HOD) from this unilateral midbrain hemorrhage. Although HOD has been associated with unilateral midbrain “rubral” tremor, it has not been described for bilateral intentional tremor. This case report illustrates how overlapping Mollaret’s triangles can explain this patient’s bilateral clinical findings. Keywords: Intentional tremor; Mollaret’s triangle; Midbrain tremor; Rubral tremor; Midbrain hemorrhage; Hypertrophic olivary degeneration Introduction Guillain-Mollaret’s triangle (GMT) is a commonly described anatomic model in association with palatal myoclonus [1]. It is also useful in the localization of tremor. The triangle consists of the dentate nucleus of the cerebellum, the red nucleus in the midbrain and the inferior olivary nucleus in the medulla. The central tegmental tract connects the red nucleus with the ipsilateral inferior olivary nucleus, while the superior cerebellar peduncle connects the dentate nucleus with OPEN ACCESS the contralateral red nucleus. The contralateral dentate nucleus and inferior olivary nucleus are *Correspondence: connected via the inferior cerebellar peduncle.