Downloaded from BOLD Systems
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Identification Key for Mosquito Species
‘Reverse’ identification key for mosquito species More and more people are getting involved in the surveillance of invasive mosquito species Species name used Synonyms Common name in the EU/EEA, not just professionals with formal training in entomology. There are many in the key taxonomic keys available for identifying mosquitoes of medical and veterinary importance, but they are almost all designed for professionally trained entomologists. Aedes aegypti Stegomyia aegypti Yellow fever mosquito The current identification key aims to provide non-specialists with a simple mosquito recog- Aedes albopictus Stegomyia albopicta Tiger mosquito nition tool for distinguishing between invasive mosquito species and native ones. On the Hulecoeteomyia japonica Asian bush or rock pool Aedes japonicus japonicus ‘female’ illustration page (p. 4) you can select the species that best resembles the specimen. On japonica mosquito the species-specific pages you will find additional information on those species that can easily be confused with that selected, so you can check these additional pages as well. Aedes koreicus Hulecoeteomyia koreica American Eastern tree hole Aedes triseriatus Ochlerotatus triseriatus This key provides the non-specialist with reference material to help recognise an invasive mosquito mosquito species and gives details on the morphology (in the species-specific pages) to help with verification and the compiling of a final list of candidates. The key displays six invasive Aedes atropalpus Georgecraigius atropalpus American rock pool mosquito mosquito species that are present in the EU/EEA or have been intercepted in the past. It also contains nine native species. The native species have been selected based on their morpho- Aedes cretinus Stegomyia cretina logical similarity with the invasive species, the likelihood of encountering them, whether they Aedes geniculatus Dahliana geniculata bite humans and how common they are. -

Zootaxa, New Records of Haemagogus
Zootaxa 1779: 65–68 (2008) ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Correspondence ZOOTAXA Copyright © 2008 · Magnolia Press ISSN 1175-5334 (online edition) New records of Haemagogus (Haemagogus) from Northern and Northeastern Brazil (Diptera: Culicidae, Aedini) JERÔNIMO ALENCAR1, FRANCISCO C. CASTRO2, HAMILTON A. O. MONTEIRO2, ORLANDO V. SILVA 2, NICOLAS DÉGALLIER3, CARLOS BRISOLA MARCONDES4*, ANTHONY E. GUIMARÃES1 1Laboratório de Diptera, Departamento de Entomologia, Instituto Oswaldo Cruz, Av. Brasil 4365, CEP: 21045-900 Manguinhos, Rio de Janeiro RJ, Brazil. 2Laboratório de Arbovírus, Instituto Evandro Chagas, Av. Almirante Barroso 492, CEP: 66090-000, Belém, PA, Brazil. 3Institut de Recherche pour le Développement (IRD-UMR182), LOCEAN-IPSL, case 100, 4 Place Jussieu, 75252 Paris Cedex 05, France 4 Departamento de Microbiologia e Parasitologia, Centro de Ciências Biológicas, Universidade Federal de Santa Catarina, 88040- 900 Florianópolis, Santa Catarina, Brazil Haemagogus (Haemagogus) is restricted mostly to the Neotropical Region, including Central America, South America and islands (Arnell, 1973). Of the 24 recognized species of this subgenus, 15 occur in South America, including the Anti- lles. However, the centre of distribution of the genus Haemagogus is Central America, where 19 of the 28 species (including four species of the subgenus Conopostegus Zavortink [1972]) occur (Arnell, 1973). Haemagogus (Hag.) includes species with great significance as vectors of Yellow Fever (YF) virus and other arbovi- rus, both experimentally (Waddell, 1949) and in the field (Vasconcelos, 2003). During entomological surveys from 1982 to 2004, the Arbovirus Laboratory of Evandro Chagas Institute obtained specimens of Haemagogus from several localities not reported in the literature. New records are listed in Table 1 and study localities shown on Figure 1. -

Diptera: Culicidae: Aedini) Into New Geographic Areas
European Mosquito Bulletin, 27 (2009), 10-17. Journal of the European Mosquito Control Association ISSN 1460-6127; w.w.w.e-m-b.org First published online 1 October 2009 Recent introductions of aedine species (Diptera: Culicidae: Aedini) into new geographic areas John F. Reinert Center for Medical, Agricultural and Veterinary Entomology (CMAVE), United States Department of Agriculture, Agricultural Research Service, 1600/1700 S.W. 23rd Drive, Gainesville, FL 32608-1067, USA, Email: [email protected]. Abstract Information on introductions to new geographic areas of species in the aedine generic-level taxa Aedimorphus, Finlaya, Georgecraigius, Halaedes, Howardina, Hulecoeteomyia, Rampamyia, Stegomyia, Tanakaius and Verrallina is provided. Key words: Aedimorphus, Finlaya, Georgecraigius atropalpus, Halaedes australis, Howardina bahamensis, Hulecoeteomyia japonica japonica, Rampamyia notoscripta, Stegomyia aegypti, Stegomyia albopicta, Tanakaius togoi, Verrallina Introduction World. Dyar (1928), however, notes that there are no nearly related species in As indicated in the series of papers on the American continent, but many such the phylogeny and classification of in the Old World, especially in Africa, mosquitoes in tribe Aedini (Reinert et and he considered that it was probably al., 2004, 2006, 2008), some aedine the African continent from which the species have been introduced into new species originated”. Christophers also geographical areas in recent times. noted that “The species is almost the Species of Aedini found outside of their only, if not the only, mosquito that, with natural ranges are listed below with their human agency, is spread around the literature citations. whole globe. But in spite of this wide zonal diffusion its distribution is very Introductions of Aedine Species to strictly limited by latitude and as far as New Areas present records go it very rarely occurs beyond latitudes of 45o N. -

Classification of Mosquitoes in Tribe Aedini 925
FORUM Classification of Mosquitoes in Tribe Aedini (Diptera: Culicidae): Paraphylyphobia, and Classification Versus Cladistic Analysis HARRY M. SAVAGE Centers for Disease Control and Prevention, P.O. Box 2087, Fort Collins, CO 80522 J. Med. Entomol. 42(6): 923Ð927 (2005) Downloaded from https://academic.oup.com/jme/article/42/6/923/886357 by guest on 29 September 2021 ABSTRACT Many mosquito species are important vectors of human and animal diseases, and others are important nuisance species. To facilitate communication and information exchange among pro- fessional groups interested in vector-borne diseases, it is essential that a stable nomenclature be maintained. For the Culicidae, easily identiÞable genera based on morphology are an asset. Major changes in generic concept, the elevation of 32 subgenera within Aedes to generic status, and changes in hundreds of species names proposed in a recent article demand consideration by all parties interested in mosquito-borne diseases. The entire approach to Aedini systematics of these authors was ßawed by an inordinate fear of paraphyletic taxa or Paraphylyphobia, and their inability to distinguish between classiÞcation and cladistic analysis. Taxonomists should refrain from making taxonomic changes based on preliminary data, and they should be very selective in assigning generic names to only the most important and well-deÞned groups of species. KEY WORDS Aedini classiÞcation, Aedes, Ochlerotatus, paraphylyphobia, mosquito classiÞcation MANY MOSQUITO SPECIES IN the tribe Aedini, a cosmo- In this communication, I brießy review taxonomic politan group represented by 11 genera and Ϸ1,239 categories in a zoological classiÞcation, the Interna- species, are important vectors of human and animal tional Code of Zoological Nomenclature (the Code), diseases, and many others are of considerable eco- the development of generic concept within the Cu- nomic importance as nuisance or pest species. -

A Multiplex PCR Assay for Six Aedini Species, Including Aedes Albopictus
A Multiplex PCR Assay for Six Aedini Species, Including Aedes albopictus Woo Jun Bang Kyungpook National University College of Natural Sciences Min Hyeok Won Kyungpook National University College of Natural Sciences Seong Tae Cho Kyungpook National University College of Natural Sciences Jihun Ryu Kyungpook National University College of Natural Sciences Kwang Shik Choi ( [email protected] ) Kyungpook National University, Daehak-ro 80, Daegu Methodology Keywords: Aedini species, Aedes albopictus, Internal transcribed spacer 2(ITS2), multiplex PCR assay Posted Date: December 15th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-127286/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Version of Record: A version of this preprint was published at Parasites & Vectors on July 28th, 2021. See the published version at https://doi.org/10.1186/s13071-021-04871-7. Page 1/8 Abstract Background Mosquitoes, as vectors of various human pathogens, are signicant drivers of serious human illness. In particular, those species in the Aedini tribe, which typically transmit dengue virus, Chikungunya fever virus, and Zika virus, are increasing their range because of climate change and international commerce. In order to prevent mosquito-borne disease, accurate mosquito species identication and monitoring are needed. The goal of this work was to develop a rapid and simple molecular diagnostic method for six morphologically similar Aedini species (Aedes avopictus, Aedes albopictus, Ochlerotatus koreicus, Ochlerotatus japonicus, Ochlerotatus togoi, and Ochlerotatus hatorii) in Korea. Methods In total, 109 samples were used in this study. The internal transcribed spacer 2 (ITS2) regions from all six species were amplied, sequenced and analyzed using Mega 6. -

The Taxonomic History of Ochlerotatus Lynch Arribálzaga, 1891 (Diptera: Culicidae)
insects Article The Taxonomic History of Ochlerotatus Lynch Arribálzaga, 1891 (Diptera: Culicidae) Lílian Ferreira de Freitas * and Lyric C. Bartholomay Department of Pathobiological Sciences, School of Veterinary Medicine, University of Wisconsin–Madison, Madison, WI 53706, USA; [email protected] * Correspondence: [email protected] Simple Summary: Mosquitoes are an extremely diverse group of aquatic insects, distributed over all continents except Antarctica. The mosquitoes belonging to the subgenus called Ochlerotatus, as understood by Reinert et al., comprise a group of several species which are endemic to the Americas, some of which are important vectors of human and animal pathogens. However, this group is characteristically undefined, i.e., no sets of characters define the group, and this presents a major challenge for the understanding of the evolutionary tree of the mosquitoes. This work underscores and contextualizes the complex taxonomic history of the group, reveals the major challenges we face in order to resolve the definition of this group, and presents a path forward for its successful revision. Abstract: A review of all taxonomic actions within the subgenus Ochlerotatus Lynch Arribálzaga, 1891 (Diptera: Culicidae) sensu Reinert et al. (2008) is provided. In particular, the complex historical taxo- nomic treatment of the type species of this group is dissected and explained in detail. Additionally, current challenges with the definition of the subgenus and its constituents are discussed, as are the requisite steps for a successful revision of the taxon. Going forward, we conclude that a taxonomic Citation: Ferreira de Freitas, L.; revision of the species should include a neotype designation for Ochlerotatus scapularis (Rondani, Bartholomay, L.C. -
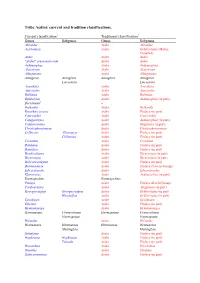
Tribe Aedini: Current and Tradition Classifications
Tribe Aedini: current and tradition classifications. Current classification1 Traditional classification2 Genus Subgenus Genus Subgenus Abraedes Aedes Abraedes Acartomyia Aedes Ochlerotatus (Mariae Complex) Aedes Aedes Aedes “Aedes” sensu auctorum Aedes Aedes Aedimorphus Aedes Aedimorphus Alanstonea Aedes Alanstonea Albuginosus Aedes Albuginosus Armigeres Armigeres Armigeres Armigeres Leicesteria Leicesteria Ayurakitia Aedes Ayurakitia Aztecaedes Aedes Aztecaedes Belkinius Aedes Belkinius Bifidistylus Aedes Aedimorphus (in part) Borichinda3 – Bothaella Aedes Bothaella Bruceharrisonius Aedes Finlaya (in part) Cancraedes Aedes Cancraedes Catageiomyia Aedes Aedimorphus (in part) Catatassomyia Aedes Stegomyia (in part) Christophersiomyia Aedes Christophersiomyia Collessius Alloeomyia Aedes Finlaya (in part) Collessius Aedes Finlaya (in part) Cornetius Aedes Cornetius Dahliana Aedes Finlaya (in part) Danielsia Aedes Finlaya (in part) Dendroskusea Aedes Diceromyia (in part) Diceromyia Aedes Diceromyia (in part) Dobrotworskyius Aedes Finlaya (in part) Downsiomyia Aedes Finlaya (Niveus Group) Edwardsaedes Aedes Edwardsaedes Elpeytonius Aedes Aedimorphus (in part) Eretmapodites Eretmapodites Finlaya Aedes Finlaya (Kochi Group) Fredwardsius Aedes Stegomyia (in part) Georgecraigius Georgecraigius Aedes Ochlerotatus (in part) Horsfallius Aedes Ochlerotatus (in part) Geoskusea Aedes Geoskusea Gilesius Aedes Finlaya (in part) Gymnometopa Aedes Gymnometopa Haemagogus Conopostegus Haemagogus Conopostegus Haemagogus Haemagogus Halaedes Aedes Halaedes Heizmannia -

Fly Times Issue 41, October 2008
FLY TIMES ISSUE 41, October, 2008 Stephen D. Gaimari, editor Plant Pest Diagnostics Branch California Department of Food & Agriculture 3294 Meadowview Road Sacramento, California 95832, USA Tel: (916) 262-1131 FAX: (916) 262-1190 Email: [email protected] Welcome to the latest issue of Fly Times! Let me first thank everyone for sending in such interesting articles – I hope you all enjoy reading it as much as I enjoyed putting it together! With that, please let me encourage all of you to consider contributing articles that may be of interest to the Diptera community. Fly Times offers a great forum to report on your research activities and to make requests for taxa being studied, as well as to report interesting observations about flies, to discuss new and improved methods, to advertise opportunities for dipterists, and to report on or announce meetings relevant to the community. This is also a great place to report on your interesting (and hopefully fruitful) collecting activities! The electronic version of the Fly Times continues to be hosted on the North American Dipterists Society website at http://www.nadsdiptera.org/News/FlyTimes/Flyhome.htm. The Diptera community would greatly appreciate your independent contributions to this newsletter. For this issue, I want to again thank all the contributors for sending me so many great articles! That said, we need even more reports on trips, collections, methods, updates, and anything else you can think of about flies, with all the associated digital images you wish to provide. Feel free to share your opinions or provide ideas on how to improve the newsletter (I am very happy to hear ways that I can enhance it!). -

Arthropod-Borne Viruses Arthropod-Borne • Rebekah C
Arthropod-Borne Viruses • Rebekah C. Kading, Aaron C Brault Beckham David and J. Arthropod-Borne Viruses The Outbreak Edition Edited by Rebekah C. Kading, Aaron C Brault and J. David Beckham Printed Edition of the Special Issue Published in Tropical Medicine and Infectious Disease www.mdpi.com/journal/tropicalmed Arthropod-Borne Viruses Arthropod-Borne Viruses: The Outbreak Edition Editors Rebekah C. Kading Aaron C Brault J. David Beckham MDPI • Basel • Beijing • Wuhan • Barcelona • Belgrade • Manchester • Tokyo • Cluj • Tianjin Editors Rebekah C. Kading Aaron C Brault Colorado State University Centers for Disease Control and Prevention USA USA J. David Beckham University of Colorado School of Medicine USA Editorial Office MDPI St. Alban-Anlage 66 4052 Basel, Switzerland This is a reprint of articles from the Special Issue published online in the open access journal Tropical Medicine and Infectious Disease (ISSN 2414-6366) (available at: https://www.mdpi.com/ journal/tropicalmed/special issues/Arthropod Borne Viruses). For citation purposes, cite each article independently as indicated on the article page online and as indicated below: LastName, A.A.; LastName, B.B.; LastName, C.C. Article Title. Journal Name Year, Article Number, Page Range. ISBN 978-3-03943-348-3 (Hbk) ISBN 978-3-03943-349-0 (PDF) c 2020 by the authors. Articles in this book are Open Access and distributed under the Creative Commons Attribution (CC BY) license, which allows users to download, copy and build upon published articles, as long as the author and publisher are properly credited, which ensures maximum dissemination and a wider impact of our publications. The book as a whole is distributed by MDPI under the terms and conditions of the Creative Commons license CC BY-NC-ND. -

Mating Systems of Blood-Feeding Flies
27 Oct 2005 13:30 AR ANRV263-EN51-18.tex XMLPublishSM(2004/02/24) P1: OKZ 10.1146/annurev.ento.51.110104.151058 Annu. Rev. Entomol. 2006. 51:413–40 doi: 10.1146/annurev.ento.51.110104.151058 Copyright c 2006 by Annual Reviews. All rights reserved First published online as a Review in Advance on September 21, 2005 MATING SYSTEMS OF BLOOD-FEEDING FLIES Boaz Yuval Department of Entomology, Hebrew University of Jerusalem, Rehovot 76100, Israel; email: [email protected] KeyWords Diptera, sexual behavior, swarm, female choice ■ Abstract The mating system of each species is a unique, dynamic suite of inter- actions between the sexes. In this review I describe these interactions in the families of flies that contain blood-feeding species. A transition from the aerial swarm, with rapid copulae and no direct female choice, to substrate-based systems with lengthy copulae and opportunities for female choice is evident at both a phylogenetic scale and within nematoceran families under specific ecological conditions. Female monogamy is associated with the former, polyandry with the latter. I suggest that the intensity of sexual selection operating on males in systems where the probability of mating is low has favored male ability to control female receptivity. Reproductive success of males is universally correlated to successful foraging for sugar or blood and (in some species and ecological conditions) to body size. Understanding the ecological basis of the mating systems of these flies will help formulate integrative, sustainable, and biologically lucid approaches for their control. INTRODUCTION Every species of sexually reproducing organism manifests a unique mating sys- tem. -

A Stable Classification of Tribe Aedini That Balances Utility with Current Knowledge of Evolutionary Relationships
RESEARCH ARTICLE Making Mosquito Taxonomy Useful: A Stable Classification of Tribe Aedini that Balances Utility with Current Knowledge of Evolutionary Relationships Richard C. Wilkerson1*, Yvonne-Marie Linton1,2,3,4, Dina M. Fonseca5, Ted R. Schultz1, Dana C. Price5, Daniel A. Strickman6 1 Department of Entomology, National Museum of Natural History, Smithsonian Institution, Washington DC, United States of America, 2 Walter Reed Biosystematics Unit, Museum Support Center, Smithsonian Institution, Suitland, Maryland, United States of America, 3 Department of Entomology, Walter Reed Army Institute of Research, Silver Spring, Maryland, United States of America, 4 Faculty of Preventative Medicine and Biometrics, Uniformed Services University of the Health Sciences, Bethesda, Maryland, United States of America, 5 Entomology Department, School of Environmental and Biological Sciences, Rutgers University, New Brunswick, New Jersey, United States of America, 6 Global Health Program, Bill and Melinda Gates Foundation, Seattle, Washington, United States of America OPEN ACCESS * [email protected] Citation: Wilkerson RC, Linton Y-M, Fonseca DM, Schultz TR, Price DC, Strickman DA (2015) Making Mosquito Taxonomy Useful: A Stable Classification of Tribe Aedini that Balances Utility with Current Abstract Knowledge of Evolutionary Relationships. PLoS ONE The tribe Aedini (Family Culicidae) contains approximately one-quarter of the known spe- 10(7): e0133602. doi:10.1371/journal.pone.0133602 cies of mosquitoes, including vectors of deadly or debilitating disease agents. This tribe Editor: Zhong-Jian Liu, The National Orchid contains the genus Aedes, which is one of the three most familiar genera of mosquitoes. Conservation Center of China; The Orchid Conservation & Research Center of Shenzhen, During the past decade, Aedini has been the focus of a series of extensive morphology- CHINA based phylogenetic studies published by Reinert, Harbach, and Kitching (RH&K). -

Diptera: Culicidae), Based on Morphological Characters
Available online at www.easletters.com Entomology and Applied Science ISSN No: 2349-2864 Letters, 2014, 1, 4:22-50 Phylogenetic relationships in the genus Psorophora Robineau-Desvoidy (Diptera: Culicidae), based on morphological characters Jonathan Liria 1 and Juan-Carlos Navarro 2 1Laboratorio Museo de Zoología, Facultad Experimental de Ciencias y Tecnología, Universidad de Carabobo, Valencia, Venezuela 2Laboratorio de Biología de Vectores, Instituto de Zoología y Ecología Tropical. Universidad Central de Venezuela. Caracas DC, Venezuela Correspondence: [email protected] (Received: 14/8/14 ) (Accepted:5/9/14) _____________________________________________________________________________________________ ABSTRACT Phylogenetic relationships in the genus Psorophora were examined based on a cladistic analysis of 66 morphological characters (fourth instar larvae, pupae, males and females) of 29 species available from the 45 species reported in the genus, representing the three subgenera. The ingroup species were: five Psorophora (10 spp total, 50%), seven Grabhamia (15 spp total, 47%) and twelve Janthinosoma (20 spp total, 60%). Two Culicini genera (Culex and Toxorhynchites), two Aedini genera (Aedes and Haemagogus), and one Mansoniini (Mansonia) were used as outgroups and sister groups respectively. A parsimony analysis using TNT resulted in 11 trees, each with 164 steps (CI = 0.66 and RI = 0.83). The analysis indicated that Aedini (Haemagogus, Aedes and Psorophora), is monophyletic group that includes Mansonia titillans (Mansoniini), suggesting natural inclusion of the Mansoniini tribe into Aedini. The genus Psorophora is monophyletic, supported by 3 synapomorphies: larvae with presence of trident-like scales in segment VIII; female with tergo and sternum VIII with rod-like structure and male genitalia with few teeth on the sternite process on segment X.