Familial Vs. Sporadic Sarcoidosis: BTNL2 Polymorphisms, Clinical
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Modulated in Intestinal Inflammation A
BTNL2, a Butyrophilin/B7-Like Molecule, Is a Negative Costimulatory Molecule Modulated in Intestinal Inflammation This information is current as Heather A. Arnett, Sabine S. Escobar, Eva Gonzalez-Suarez, of September 28, 2021. Alison L. Budelsky, Lori A. Steffen, Norman Boiani, Ming Zhang, Gerald Siu, Avery W. Brewer and Joanne L. Viney J Immunol 2007; 178:1523-1533; ; doi: 10.4049/jimmunol.178.3.1523 http://www.jimmunol.org/content/178/3/1523 Downloaded from References This article cites 40 articles, 12 of which you can access for free at: http://www.jimmunol.org/content/178/3/1523.full#ref-list-1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication by guest on September 28, 2021 *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2007 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology BTNL2, a Butyrophilin/B7-Like Molecule, Is a Negative Costimulatory Molecule Modulated in Intestinal Inflammation Heather A. Arnett,1* Sabine S. -
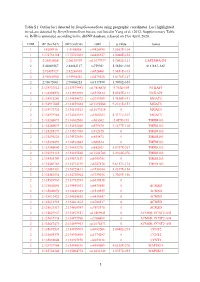
1 Table S1. Outlier Loci Detected by Deepgenomescan Using
Table S1. Outlier loci detected by DeepGenomeScan using geographic coordinates. Loci highlighted in red are detected by DeepGenomeScan but are not listed in Yang et al. (2012; Supplementary Table 4). RsID is annotated according to the dbSNP database released on 21st April, 2020. CHR BP (Grch37) BP (Grch38) rsID p.value Genes 1 1:4208918 1:4148858 rs9426495 3.8582E-104 1 1:175738168 1:175769032 rs6425357 2.0088E-105 2 2:20310668 2:20110907 rs11679737 1.2002E-111 LAPTM4A-DT 2 2:40289267 2:40062127 rs759361 5.5658E-104 SLC8A1-AS1 2 2:82495127 2:82268003 rs6726401 3.9451E-103 2 2:96660300 2:95994552 rs2579520 1.8176E-127 2 2:96672001 2:96006253 rs1917890 3.7698E-104 2 2:134333012 2:133575441 rs17816830 9.702E-105 NCKAP5 2 2:134350570 2:133592999 rs6715224 5.8745E-111 NCKAP5 2 2:134912243 2:134154672 rs2139309 1.7418E-191 MGAT5 2 2:134917005 2:134159434 rs11692586 9.2121E-151 MGAT5 2 2:134972732 2:134215161 rs11679218 0 MGAT5 2 2:134979966 2:134222395 rs1965183 5.3171E-107 MGAT5 2 2:135260071 2:134502500 rs503562 2.057E-153 TMEM163 2 2:135280039 2:134522468 rs579670 3.1477E-118 TMEM163 2 2:135285279 2:134527708 rs512375 0 TMEM163 2 2:135290221 2:134532650 rs655472 0 TMEM163 2 2:135290453 2:134532882 rs666614 0 TMEM163 2 2:135340840 2:134583270 rs842361 5.6717E-237 TMEM163 2 2:135393110 2:134635540 rs11684785 2.2164E-276 TMEM163 2 2:135430709 2:134673139 rs6745983 0 TMEM163 2 2:135469769 2:134712199 rs6747870 9.6117E-114 TMEM163 2 2:135483381 2:134725811 rs3739034 5.0357E-154 2 2:135483534 2:134725964 rs3739036 3.7269E-156 2 2:135539967 2:134782397 -

Next Generation Exome Sequencing of Paediatric Inflammatory Bowel Disease Patients Identifies Rare and Novel Variants in Candida
Gut Online First, published on April 28, 2012 as 10.1136/gutjnl-2011-301833 Inflammatory bowel disease ORIGINAL ARTICLE Gut: first published as 10.1136/gutjnl-2011-301833 on 28 April 2012. Downloaded from Next generation exome sequencing of paediatric inflammatory bowel disease patients identifies rare and novel variants in candidate genes Katja Christodoulou,1 Anthony E Wiskin,2 Jane Gibson,1 William Tapper,1 Claire Willis,2 Nadeem A Afzal,3 Rosanna Upstill-Goddard,1 John W Holloway,4 Michael A Simpson,5 R Mark Beattie,3 Andrew Collins,1 Sarah Ennis1 < Additional materials are ABSTRACT published online only. To view Background Multiple genes have been implicated by Significance of this study these files please visit the association studies in altering inflammatory bowel journal online (http://gut.bmj. com/content/early/recent). disease (IBD) predisposition. Paediatric patients often What is already known on this subject? manifest more extensive disease and a particularly < For numbered affiliations see Genome-wide association studies have impli- end of article. severe disease course. It is likely that genetic cated numerous candidate genes for inflamma- predisposition plays a more substantial role in this group. tory bowel disease (IBD), but evidence of Correspondence to Objective To identify the spectrum of rare and novel causality for specific variants is largely absent. Dr Sarah Ennis, Genetic variation in known IBD susceptibility genes using exome Furthermore, by design, genome-wide associa- Epidemiology and Genomic sequencing analysis in eight individual cases of childhood Informatics Group, Human tion studies are limited to the study of Genetics, Faculty of Medicine, onset severe disease. -

Familial Vs. Sporadic Sarcoidosis: BTNL2 Polymorphisms, Clinical
Familial vs. sporadic sarcoidosis: BTNL2 polymorphisms, clinical presentations, and outcomes in a French cohort Yves Pacheco, Alain Calender, Dominique Israël-Biet, Pascal Roy, Serge Lebecque, Vincent Cottin, Diane Bouvry, Hilario Nunes, Pascal Sève, Laurent Pérard, et al. To cite this version: Yves Pacheco, Alain Calender, Dominique Israël-Biet, Pascal Roy, Serge Lebecque, et al.. Familial vs. sporadic sarcoidosis: BTNL2 polymorphisms, clinical presentations, and outcomes in a French cohort. Orphanet Journal of Rare Diseases, BioMed Central, 2016, 11 (1), 10.1186/s13023-016-0546-4. hal- 01595465 HAL Id: hal-01595465 https://hal.archives-ouvertes.fr/hal-01595465 Submitted on 26 Sep 2017 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - ShareAlike| 4.0 International License Pacheco et al. Orphanet Journal of Rare Diseases (2016) 11:165 DOI 10.1186/s13023-016-0546-4 RESEARCH Open Access Familial vs. sporadic sarcoidosis: BTNL2 polymorphisms, clinical presentations, and outcomes in a French cohort Yves Pacheco1,11*, Alain Calender2, Dominique Israël-Biet3, Pascal Roy4, Serge Lebecque5, Vincent Cottin6, Diane Bouvry7, Hilario Nunes7, Pascal Sève8, Laurent Pérard9, Gilles Devouassoux8, Nathalie Freymond1, Chahira Khouatra6, Benoît Wallaert10, Raphaelle Lamy2, Mad-Hélénie Elsensohn4, Claire Bardel4, Dominique Valeyre7 and GSF group Abstract Background: The occurrence of familial forms of sarcoidosis (OMIM 181100) suggests a genetic predisposition. -

2027.Full.Pdf
Butyrophilin-like 2 Modulates B7 Costimulation To Induce Foxp3 Expression and Regulatory T Cell Development in Mature T Cells This information is current as of September 28, 2021. Ryan M. Swanson, Marc A. Gavin, Sabine S. Escobar, James B. Rottman, Brian P. Lipsky, Shishir Dube, Li Li, Jeannette Bigler, Martin Wolfson, Heather A. Arnett and Joanne L. Viney J Immunol 2013; 190:2027-2035; Prepublished online 28 Downloaded from January 2013; doi: 10.4049/jimmunol.1201760 http://www.jimmunol.org/content/190/5/2027 http://www.jimmunol.org/ Supplementary http://www.jimmunol.org/content/suppl/2013/01/28/jimmunol.120176 Material 0.DC1 References This article cites 40 articles, 18 of which you can access for free at: http://www.jimmunol.org/content/190/5/2027.full#ref-list-1 by guest on September 28, 2021 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. -

Functions to Inhibit T Cell Activation BTNL2, a Butyrophilin-Like
BTNL2, a Butyrophilin-Like Molecule That Functions to Inhibit T Cell Activation Thang Nguyen, Xikui K. Liu, Yongliang Zhang and Chen Dong This information is current as of October 2, 2021. J Immunol 2006; 176:7354-7360; ; doi: 10.4049/jimmunol.176.12.7354 http://www.jimmunol.org/content/176/12/7354 Downloaded from References This article cites 40 articles, 10 of which you can access for free at: http://www.jimmunol.org/content/176/12/7354.full#ref-list-1 Why The JI? Submit online. http://www.jimmunol.org/ • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average by guest on October 2, 2021 Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2006 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology BTNL2, a Butyrophilin-Like Molecule That Functions to Inhibit T Cell Activation1 Thang Nguyen,2* Xikui K. Liu,2† Yongliang Zhang,† and Chen Dong3† B7 family members regulate T cell activation and tolerance. Although butyrophilin proteins share sequence homology with the B7 molecules, it is unclear whether they have any function in immune responses. -

FARE2017 WINNERS Sorted by Institute/Center
FARE2017 WINNERS Sorted By Institute/Center NIH Clinical Center (CC) Zachary Lerner Postdoctoral FellowPostdoctoral Fellow Clinical and Translational Research A Robotic Exoskeleton to Treat Crouch Gait from Cerebral Palsy: Design and Initial Clinical Evaluation Cerebral palsy (CP) is the most prevalent childhood physical disability and adversely affects walking and other motor abilities. Crouch gait, a pathological walking pattern characterized by excessive knee flexion, is one of the most common gait disorders observed in children with CP. Effective treatment of crouch during childhood is critical to maintain mobility into adulthood, yet current interventions do not adequately eliminate or alleviate crouch in most patients. Wearable robotic exoskeletons have the potential to improve crouch gait by providing on demand assistance during walking. However, no exoskeletons suitable to treat children are commercially available, and no evidence exists regarding the feasibility or efficacy of utilizing motorized assistance to alleviate knee flexion from crouch gait. Enhanced knowledge of neuromuscular adaptations to powered assistance in children with crouch gait is needed to optimize this treatment approach. To meet these needs, we developed the first lower- extremity exoskeleton intended to treat crouch gait by providing powered knee extension assistance at different phases of the gait cycle. We evaluated the effects of powered knee extension assistance on knee motion, and knee flexor and extensor muscle activity in four children with crouch gait from CP. The exoskeleton was effective in reducing crouch in three of the four participants compared to their baseline gait pattern. The reduction in crouch was clinically significant (greater than 10 degrees) in two of the participants. -

Functional Characterization of Potential New Agents for Cancer Immunotherapy
Functional Characterization of Potential New Agents for Cancer Immunotherapy The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:39010116 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA Functional Characterization of Potential New Agents for Cancer Immunotherapy Xiaoxu Wang A Thesis Submitted to the Faculty of The Harvard Medical School in Partial Fulfillment of the Requirements for the Degree of Master of Medical Sciences in Immunology Harvard University Boston, Massachusetts. May, 2018 Thesis Advisor: Dr. Gordon Freeman Xiaoxu Wang Functional Characterization of Potential New Agents for Cancer Immunotherapy Abstract PD-1/PD-L1 checkpoint blockade has demonstrated promise in a variety of malignancies. The blockade of T cell inhibitory molecules PD-1 and PD-L1 can rejuvenate T cells and improve tumor cell killing. However, the overall response rate of PD-1/PD-L1 immunotherapy is only about 20-30%. One promising method to improve cancer immunotherapy is combinatory immunotherapies that target new receptors/ligands regulating T cell activity, so the functional characterization of these new receptors/ligands is urgent. Due to the functional importance of the B7 superfamily in T cell activation, I studied T cell regulating functions of two poorly characterized B7 superfamily members, IgC domain-truncated CD86 and butyrophilin-like 2 (BTNL2). -

Detection of BTNL2 Gene Mutation (Rs2076530 Allele) in Iranian
icine- O ed pe M n l A Vazifehmand et al., Intern Med 2017, 7:3 a c n c r e e s t DOI: 10.4172/2165-8048.1000239 s n I Internal Medicine: Open Access ISSN: 2165-8048 Research Article Open Access Detection of BTNL2 Gene Mutation (rs2076530 Allele) in Iranian Sarcoidosis Patients: A clinical and Genetic Study Reza Vazifehmand1, Tina Saber2, Hamid Reza Khorram Khorshid3, Dhuha Saeed Ali4, Foroozandeh monem homaie5 and Sassan Saber6* 1Young Researchers and elite Club, Islamic Azad University- Rasht campus, Iran 2Beckman Laser Institute and Medical Clinic, University of California, Irvine, Ca, USA 3Genetic Research Centre, University of Social Welfare and Rehabilitation Sciences, Tehran, Iran 4Halal Products Research Institute, Universiti Putra Malaysia. Serdang,Selangor-Malaysia 5Departments of Biochemistry, Science and Research University-Fars Branch-Shiraz-Iran 6North Karegar Ave. Shariati Hospital, Faculty of Medicine, Tehran University of Medical Science-Tehran, Iran *Corresponding Author: Sassan Saber, North Karegar Ave. Shariati Hospital, Faculty of Medicine Tehran University of Medical Science, Tel: 00982188220000; E-mail: [email protected] Received date: October 24, 2016; Accepted date: May 05, 2017; Published date: May 15, 2017 Copyright: © 2017 Vazifehmand R, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Abstract Background/Aims: Sarcoidosis is a multiorgan granulomatous inflammatory disease of an unknown cause, probably due to inappropriate T-cell response. Mutation in BTNL2 gene (Butirophylin-like2) which is one of the most important genes in MHC II (complex tissue incompatibility) group is related to sarcoidosis. -

Leveraging Models of Cell Regulation and GWAS Data in Integrative Network-Based Association Studies
PERSPECTIVE Leveraging models of cell regulation and GWAS data in integrative network-based association studies Andrea Califano1–3,11, Atul J Butte4,5, Stephen Friend6, Trey Ideker7–9 & Eric Schadt10,11 Over the last decade, the genome-wide study of both heritable and straightforward: within the space of all possible genetic and epigenetic somatic human variability has gone from a theoretical concept to a variants, those contributing to a specific trait or disease likely have broadly implemented, practical reality, covering the entire spectrum some coalescent properties, allowing their effect to be functionally of human disease. Although several findings have emerged from these canalized via the cell communication and cell regulatory machin- studies1, the results of genome-wide association studies (GWAS) have ery that allows distinct cells to interact and regulates their behavior. been mostly sobering. For instance, although several genes showing Notably, contrary to random networks, whose output is essentially medium-to-high penetrance within heritable traits were identified by unconstrained, regulatory networks produced by adaptation to spe- these approaches, the majority of heritable genetic risk factors for most cific fitness landscapes are optimized to produce only a finite number common diseases remain elusive2–7. Additionally, due to impractical of well-defined outcomes as a function of a very large number of exog- requirements for cohort size8 and lack of methodologies to maximize enous and endogenous signals. Thus, if a comprehensive and accurate power for such detections, few epistatic interactions and low-pen- map of all intra- and intercellular molecular interactions were avail- etrance variants have been identified9. At the opposite end of the able, then genetic and epigenetic events implicated in a specific trait or germline versus somatic event spectrum, considering that tumor cells disease should cluster in subnetworks of closely interacting genes. -

Use of Deep-Learning Genomics to Discriminate Healthy Individuals from Those with Alzheimer’S Disease Or Mild Cognitive Impairment
Hindawi Behavioural Neurology Volume 2021, Article ID 3359103, 15 pages https://doi.org/10.1155/2021/3359103 Research Article Use of Deep-Learning Genomics to Discriminate Healthy Individuals from Those with Alzheimer’s Disease or Mild Cognitive Impairment Lanlan Li,1 Yeying Yang,2 Qi Zhang,1 Jiao Wang,3 Jiehui Jiang ,1 and Alzheimer’s Disease Neuroimaging Initiative4 1Institute of Biomedical Engineering, School of Communication and Information Engineering, Shanghai University, Shanghai 200444, China 2LongHua Hospital, Shanghai University of Traditional Chinese Medicine, Shanghai 200032, China 3School of Life Science, Shanghai University, Shanghai 200444, China 4Indiana University School of Medicine, Indianapolis, IN 46202, USA Correspondence should be addressed to Jiehui Jiang; [email protected] Received 14 May 2021; Accepted 11 June 2021; Published 15 July 2021 Academic Editor: Muh-Shi Lin Copyright © 2021 Lanlan Li et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Objectives. Alzheimer’s disease (AD) is the most prevalent neurodegenerative disorder and the most common form of dementia in the elderly. Certain genes have been identified as important clinical risk factors for AD, and technological advances in genomic research, such as genome-wide association studies (GWAS), allow for analysis of polymorphisms and have been widely applied to studies of AD. However, shortcomings of GWAS include sensitivity to sample size and hereditary deletions, which result in low classification and predictive accuracy. Therefore, this paper proposes a novel deep-learning genomics approach and applies it to multitasking classification of AD progression, with the goal of identifying novel genetic biomarkers overlooked by traditional GWAS analysis. -

A Meta-Analysis of the Effects of High-LET Ionizing Radiations in Human Gene Expression
Supplementary Materials A Meta-Analysis of the Effects of High-LET Ionizing Radiations in Human Gene Expression Table S1. Statistically significant DEGs (Adj. p-value < 0.01) derived from meta-analysis for samples irradiated with high doses of HZE particles, collected 6-24 h post-IR not common with any other meta- analysis group. This meta-analysis group consists of 3 DEG lists obtained from DGEA, using a total of 11 control and 11 irradiated samples [Data Series: E-MTAB-5761 and E-MTAB-5754]. Ensembl ID Gene Symbol Gene Description Up-Regulated Genes ↑ (2425) ENSG00000000938 FGR FGR proto-oncogene, Src family tyrosine kinase ENSG00000001036 FUCA2 alpha-L-fucosidase 2 ENSG00000001084 GCLC glutamate-cysteine ligase catalytic subunit ENSG00000001631 KRIT1 KRIT1 ankyrin repeat containing ENSG00000002079 MYH16 myosin heavy chain 16 pseudogene ENSG00000002587 HS3ST1 heparan sulfate-glucosamine 3-sulfotransferase 1 ENSG00000003056 M6PR mannose-6-phosphate receptor, cation dependent ENSG00000004059 ARF5 ADP ribosylation factor 5 ENSG00000004777 ARHGAP33 Rho GTPase activating protein 33 ENSG00000004799 PDK4 pyruvate dehydrogenase kinase 4 ENSG00000004848 ARX aristaless related homeobox ENSG00000005022 SLC25A5 solute carrier family 25 member 5 ENSG00000005108 THSD7A thrombospondin type 1 domain containing 7A ENSG00000005194 CIAPIN1 cytokine induced apoptosis inhibitor 1 ENSG00000005381 MPO myeloperoxidase ENSG00000005486 RHBDD2 rhomboid domain containing 2 ENSG00000005884 ITGA3 integrin subunit alpha 3 ENSG00000006016 CRLF1 cytokine receptor like