Use of Deep-Learning Genomics to Discriminate Healthy Individuals from Those with Alzheimer’S Disease Or Mild Cognitive Impairment
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Modulated in Intestinal Inflammation A
BTNL2, a Butyrophilin/B7-Like Molecule, Is a Negative Costimulatory Molecule Modulated in Intestinal Inflammation This information is current as Heather A. Arnett, Sabine S. Escobar, Eva Gonzalez-Suarez, of September 28, 2021. Alison L. Budelsky, Lori A. Steffen, Norman Boiani, Ming Zhang, Gerald Siu, Avery W. Brewer and Joanne L. Viney J Immunol 2007; 178:1523-1533; ; doi: 10.4049/jimmunol.178.3.1523 http://www.jimmunol.org/content/178/3/1523 Downloaded from References This article cites 40 articles, 12 of which you can access for free at: http://www.jimmunol.org/content/178/3/1523.full#ref-list-1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication by guest on September 28, 2021 *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2007 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology BTNL2, a Butyrophilin/B7-Like Molecule, Is a Negative Costimulatory Molecule Modulated in Intestinal Inflammation Heather A. Arnett,1* Sabine S. -

Gene Polymorphisms of Serotonin Receptors and Drug-Induced Hyperprolactinemia in Patients with Schizophrenia
Poster number: P.3.b.037 Gene polymorphisms of serotonin receptors and drug-induced hyperprolactinemia in patients with schizophrenia Diana Z. Оsmanova1, Anastasia S. Boiko1, Olga Yu. Fedorenko1, Ivan V. Pozhidaev1, M.B. Freidin1 Elena G. Kornetova1, Svetlana A. Ivanova1 , Bob Wilffert2, Anton J.M. Loonen2 1. Mental Health Research Institute, Tomsk National Research Medical Center, Russian Academy of Sciences, Tomsk, Russia 2. Department of Pharmacy, University of Groningen, Groningen, The Netherlands BACKGROUND RESULTS Antipsychotic drug-induced hyperprolactinemia is an All patients with schizophrenia were divided into two increasingly prevalent problem in current psychiatric practice and groups: those with and without hyperprolactinemia. Patients responsible for troublesome side effects like loss of libido and from both groups were genotyped for HTR1A variants: rs6295, impotence. The chance to develop hyperprolactinemia depends rs1364043, rs10042486, rs1800042, rs749099; for HTR1B: upon the pharmacological properties of antipsychotic medication rs6298, rs6296, rs130058; for HTR2A: rs6311, rs6313, rs6314, used, of its dosage and treatment duration, as well as from the rs7997012, rs1928040, rs9316233, rs2224721, rs6312; for genetic make-up and other characteristics which determine the HTR2C: rs6318, rs5946189, rs569959, rs17326429, rs4911871, individual sensitivity of the individual patient. rs3813929, rs1801412, rs12858300; for HTR3A: rs1062613, Second generation antipsychotics are (often) more potent rs33940208, rs1176713; for HTR3B: -

Pharmacogenetics of Antidepressants, a Review Of
al Depres ic sio lin n C Reyes-Barron et al., Clin Depress 2016, 2:2 Clinical Depression Research Article Article OpenOpen Access Access Pharmacogenetics of Antidepressants, A Review of Significant Genetic Variants in Different Populations Cynthia Reyes-Barron1, Silvina Tonarelli1, Andrew Delozier1, David F. Briones1, Brenda B. Su2, Lewis P. Rubin1 and Chun Xu1* 1Texas Tech University Health Sciences Centre, Paul L. Foster School of Medicine 2College of Medicine and Health Sciences, UAE University, Department of Internal Medicine Abstract Major depressive disorder is a highly prevalent disease that is challenging to treat, often requiring medication and dose adjustments. Genetic factors play an important role in psychotropic medication responses. However, the translation of pharmacogenetics findings to clinical recommendations with regards to antidepressant responses is still in its early stages. We reviewed recent primary research articles, meta-analyses, and reviews on the pharmacogenetics of antidepressant treatment for major depressive disorder in different populations. We identified eight genes with likely associations with treatment responses and summarized genetic variants most likely to influence treatment responses. We determined the frequency of these variants in Caucasian, Asian, Hispanic, and African American populations. The genes are related to functions in drug metabolism, transport, signalling, stress response, and neuroplasticity. Clinical recommendations already exist for CYP2D6 and CYP2C19 cytochrome P450 drug metabolism genes. The other genes are: ABCB1 with single nucleotide polymorphisms (SNPs) rs2032583 and rs2235015; FKBP5 with SNPs rs1360780, rs3800373, and rs4713916; GNB3 with SNP rs5443; BDNF with SNP rs6265; HTR2A with SNPs rs7997012 and rs6313; and SLC6A4 with polymorphisms 5-HTTLPR and STin2. There is significant variability of the frequencies of these polymorphisms in the different populations we reviewed. -

A Serotonin Receptor with a Possible Role in Joint Diseases
Anders Kling 5-HT2 A – a serotonin receptor with a possible role in joint diseases role with a possible receptor – a serotonin 5-HT2A – a serotonin receptor with a possible role in joint diseases Anders Kling Umeå University 2013 Umeå University Department of Pharmacology and Clinical Neuroscience New Serie 1547 Department of Pharmacology and Clinical Neurosciences Umeå University ISSN: 0346-6612 Umeå University, Sweden 2013 SE-901 87 Umeå, Sweden ISBN 978-91-7459-549-9 5-HT2A – a serotonin receptor with a possible role in joint diseases Anders Kling Institutionen för farmakologi och klinisk neurovetenskap, Klinisk farmakologi/ Department of Pharmacology and Clinical Neuroscience, Clinical Pharmacology Umeå universitet/ Umeå University Umeå 2013 Responsible publisher under swedish law: the Dean of the Medical Faculty This work is protected by the Swedish Copyright Legislation (Act 1960:729) ISBN: 978-91-7459-549-9 ISSN: 0346-6612 New series No: 1547 Elektronisk version tillgänglig på http://umu.diva-portal.org/ Tryck/Printed by: Print och Media, Umeå universitet Umeå, Sweden 2013 Innehåll/Table of Contents Innehåll/Table of Contents i Abstract iv Abbreviations vi List of studies viii Populärvetenskaplig sammanfattning ix 5-HT2A – en serotoninreceptor med möjlig betydelse för ledsjukdomar ix Introduction 1 The serotonin system 1 Serotonin 1 Serotonin receptors 2 The serotonin system and platelets 2 Serotonin receptor 5-HT2A 3 Localisation/expression of 5-HT2A receptors 3 Functions of the 5-HT2A receptor 4 Regulation of the 5-HT2A receptor -

Molecular Genetics of Human Personality Traits for Psychiatric, Behav- Ioral, and Substance-Related Disorders Eugene Lin*,1 and Po See Chen*,2,3
The Open Translational Medicine Journal, 2009, 1, 1-8 1 Open Access Molecular Genetics of Human Personality Traits for Psychiatric, Behav- ioral, and Substance-Related Disorders Eugene Lin*,1 and Po See Chen*,2,3 1Vita Genomics, Inc., 7 Fl., No. 6, Sec. 1, Jung-Shing Road, Wugu Shiang, Taipei, Taiwan 2Department of Psychiatry, National Cheng Kung University, Tainan, Taiwan 3National Cheng Kung University Hospital and Dou-Liou Branch, Taiwan Abstract: The investigation of personality genetics had received much attention since the three seminal reports showing an association between genes and personality traits in the general population. Accumulating evidences suggested that per- sonality traits have significant genetic components. Although currently available data are not enough for proof, more and more genetic variants associated with personality traits are being discovered. In this paper, we review related studies of gene polymorphisms and human personality traits for psychiatric, behavioral, and substance-related disorders. First, we briefly describe the commonly-used self-reported temperament measures that define personality dimensions. Then, we summarize the characteristics of the candidate genes for personality traits, and investigate gene variants which have been suggested to be linked with personality traits for individuals with psychiatric, behavioral, and substance-related disorders. Keywords: Molecular genetics, personality, psychiatric disorders, temperament measures. 1. INTRODUCTION and 5-HTTLPR but not other anxiety-related personality traits [14,15]. The investigation of personality genetics had received much attention since the three seminal reports [1-3] in 1996 We reviewed related studies of gene polymorphisms and showing an association between genes and personality traits human personality traits for psychiatric, behavioral, and sub- in the general population. -
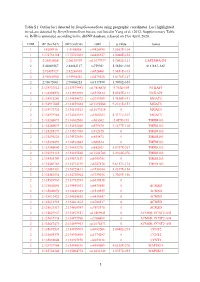
1 Table S1. Outlier Loci Detected by Deepgenomescan Using
Table S1. Outlier loci detected by DeepGenomeScan using geographic coordinates. Loci highlighted in red are detected by DeepGenomeScan but are not listed in Yang et al. (2012; Supplementary Table 4). RsID is annotated according to the dbSNP database released on 21st April, 2020. CHR BP (Grch37) BP (Grch38) rsID p.value Genes 1 1:4208918 1:4148858 rs9426495 3.8582E-104 1 1:175738168 1:175769032 rs6425357 2.0088E-105 2 2:20310668 2:20110907 rs11679737 1.2002E-111 LAPTM4A-DT 2 2:40289267 2:40062127 rs759361 5.5658E-104 SLC8A1-AS1 2 2:82495127 2:82268003 rs6726401 3.9451E-103 2 2:96660300 2:95994552 rs2579520 1.8176E-127 2 2:96672001 2:96006253 rs1917890 3.7698E-104 2 2:134333012 2:133575441 rs17816830 9.702E-105 NCKAP5 2 2:134350570 2:133592999 rs6715224 5.8745E-111 NCKAP5 2 2:134912243 2:134154672 rs2139309 1.7418E-191 MGAT5 2 2:134917005 2:134159434 rs11692586 9.2121E-151 MGAT5 2 2:134972732 2:134215161 rs11679218 0 MGAT5 2 2:134979966 2:134222395 rs1965183 5.3171E-107 MGAT5 2 2:135260071 2:134502500 rs503562 2.057E-153 TMEM163 2 2:135280039 2:134522468 rs579670 3.1477E-118 TMEM163 2 2:135285279 2:134527708 rs512375 0 TMEM163 2 2:135290221 2:134532650 rs655472 0 TMEM163 2 2:135290453 2:134532882 rs666614 0 TMEM163 2 2:135340840 2:134583270 rs842361 5.6717E-237 TMEM163 2 2:135393110 2:134635540 rs11684785 2.2164E-276 TMEM163 2 2:135430709 2:134673139 rs6745983 0 TMEM163 2 2:135469769 2:134712199 rs6747870 9.6117E-114 TMEM163 2 2:135483381 2:134725811 rs3739034 5.0357E-154 2 2:135483534 2:134725964 rs3739036 3.7269E-156 2 2:135539967 2:134782397 -

Next Generation Exome Sequencing of Paediatric Inflammatory Bowel Disease Patients Identifies Rare and Novel Variants in Candida
Gut Online First, published on April 28, 2012 as 10.1136/gutjnl-2011-301833 Inflammatory bowel disease ORIGINAL ARTICLE Gut: first published as 10.1136/gutjnl-2011-301833 on 28 April 2012. Downloaded from Next generation exome sequencing of paediatric inflammatory bowel disease patients identifies rare and novel variants in candidate genes Katja Christodoulou,1 Anthony E Wiskin,2 Jane Gibson,1 William Tapper,1 Claire Willis,2 Nadeem A Afzal,3 Rosanna Upstill-Goddard,1 John W Holloway,4 Michael A Simpson,5 R Mark Beattie,3 Andrew Collins,1 Sarah Ennis1 < Additional materials are ABSTRACT published online only. To view Background Multiple genes have been implicated by Significance of this study these files please visit the association studies in altering inflammatory bowel journal online (http://gut.bmj. com/content/early/recent). disease (IBD) predisposition. Paediatric patients often What is already known on this subject? manifest more extensive disease and a particularly < For numbered affiliations see Genome-wide association studies have impli- end of article. severe disease course. It is likely that genetic cated numerous candidate genes for inflamma- predisposition plays a more substantial role in this group. tory bowel disease (IBD), but evidence of Correspondence to Objective To identify the spectrum of rare and novel causality for specific variants is largely absent. Dr Sarah Ennis, Genetic variation in known IBD susceptibility genes using exome Furthermore, by design, genome-wide associa- Epidemiology and Genomic sequencing analysis in eight individual cases of childhood Informatics Group, Human tion studies are limited to the study of Genetics, Faculty of Medicine, onset severe disease. -

Familial Vs. Sporadic Sarcoidosis: BTNL2 Polymorphisms, Clinical
Familial vs. sporadic sarcoidosis: BTNL2 polymorphisms, clinical presentations, and outcomes in a French cohort Yves Pacheco, Alain Calender, Dominique Israël-Biet, Pascal Roy, Serge Lebecque, Vincent Cottin, Diane Bouvry, Hilario Nunes, Pascal Sève, Laurent Pérard, et al. To cite this version: Yves Pacheco, Alain Calender, Dominique Israël-Biet, Pascal Roy, Serge Lebecque, et al.. Familial vs. sporadic sarcoidosis: BTNL2 polymorphisms, clinical presentations, and outcomes in a French cohort. Orphanet Journal of Rare Diseases, BioMed Central, 2016, 11 (1), 10.1186/s13023-016-0546-4. hal- 01595465 HAL Id: hal-01595465 https://hal.archives-ouvertes.fr/hal-01595465 Submitted on 26 Sep 2017 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - ShareAlike| 4.0 International License Pacheco et al. Orphanet Journal of Rare Diseases (2016) 11:165 DOI 10.1186/s13023-016-0546-4 RESEARCH Open Access Familial vs. sporadic sarcoidosis: BTNL2 polymorphisms, clinical presentations, and outcomes in a French cohort Yves Pacheco1,11*, Alain Calender2, Dominique Israël-Biet3, Pascal Roy4, Serge Lebecque5, Vincent Cottin6, Diane Bouvry7, Hilario Nunes7, Pascal Sève8, Laurent Pérard9, Gilles Devouassoux8, Nathalie Freymond1, Chahira Khouatra6, Benoît Wallaert10, Raphaelle Lamy2, Mad-Hélénie Elsensohn4, Claire Bardel4, Dominique Valeyre7 and GSF group Abstract Background: The occurrence of familial forms of sarcoidosis (OMIM 181100) suggests a genetic predisposition. -

Case–Control Association Study of 59 Candidate Genes Reveals the DRD2
Journal of Human Genetics (2009) 54, 98–107 & 2009 The Japan Society of Human Genetics All rights reserved 1434-5161/09 $32.00 www.nature.com/jhg ORIGINAL ARTICLE Case–control association study of 59 candidate genes reveals the DRD2 SNP rs6277 (C957T) as the only susceptibility factor for schizophrenia in the Bulgarian population Elitza T Betcheva1, Taisei Mushiroda2, Atsushi Takahashi3, Michiaki Kubo4, Sena K Karachanak5, Irina T Zaharieva5, Radoslava V Vazharova5, Ivanka I Dimova5, Vihra K Milanova6, Todor Tolev7, George Kirov8, Michael J Owen8, Michael C O’Donovan8, Naoyuki Kamatani3, Yusuke Nakamura1,9 and Draga I Toncheva5 The development of molecular psychiatry in the last few decades identified a number of candidate genes that could be associated with schizophrenia. A great number of studies often result with controversial and non-conclusive outputs. However, it was determined that each of the implicated candidates would independently have a minor effect on the susceptibility to that disease. Herein we report results from our replication study for association using 255 Bulgarian patients with schizophrenia and schizoaffective disorder and 556 Bulgarian healthy controls. We have selected from the literatures 202 single nucleotide polymorphisms (SNPs) in 59 candidate genes, which previously were implicated in disease susceptibility, and we have genotyped them. Of the 183 SNPs successfully genotyped, only 1 SNP, rs6277 (C957T) in the DRD2 gene (P¼0.0010, odds ratio¼1.76), was considered to be significantly associated with schizophrenia after the replication study using independent sample sets. Our findings support one of the most widely considered hypotheses for schizophrenia etiology, the dopaminergic hypothesis. -

2027.Full.Pdf
Butyrophilin-like 2 Modulates B7 Costimulation To Induce Foxp3 Expression and Regulatory T Cell Development in Mature T Cells This information is current as of September 28, 2021. Ryan M. Swanson, Marc A. Gavin, Sabine S. Escobar, James B. Rottman, Brian P. Lipsky, Shishir Dube, Li Li, Jeannette Bigler, Martin Wolfson, Heather A. Arnett and Joanne L. Viney J Immunol 2013; 190:2027-2035; Prepublished online 28 Downloaded from January 2013; doi: 10.4049/jimmunol.1201760 http://www.jimmunol.org/content/190/5/2027 http://www.jimmunol.org/ Supplementary http://www.jimmunol.org/content/suppl/2013/01/28/jimmunol.120176 Material 0.DC1 References This article cites 40 articles, 18 of which you can access for free at: http://www.jimmunol.org/content/190/5/2027.full#ref-list-1 by guest on September 28, 2021 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. -

Association of BDNF, HTR2A, TPH1, SLC6A4, and COMT
Braz J Psychiatry. 2019 xxx-xxx;00(00):000-000 doi:10.1590/1516-4446-2019-0620 Brazilian Psychiatric Association 00000000-0002-7316-1185 ORIGINAL ARTICLE Association of BDNF, HTR2A, TPH1, SLC6A4, and COMT polymorphisms with tDCS and escitalopram efficacy: ancillary analysis of a double-blind, placebo-controlled trial Andre R. Brunoni0000-0000-0000-0000 ,1,2 Angel Carracedo,3 Olalla M. Amigo,3 Ana L. Pellicer,3 Leda Talib,2 Andre F. Carvalho0000-0000-0000-0000 ,4 Paulo A. Lotufo,1 Isabela M. Bensen˜ or,1 Wagner Gattaz,2 Carolina Cappi5 1Departamento de Medicina Interna, Faculdade de Medicina, Universidade de Sa˜o Paulo (USP), Sa˜o Paulo, SP, Brazil. 2Laborato´rio de Neurocieˆncias (LIM-27) and Instituto Nacional de Biomarcadores em Psiquiatria (INBION), Departamento e Instituto de Psiquiatria, Faculdade de Medicina, USP, Sa˜o Paulo, SP, Brazil. 3Grupo de Medicina Xeno´mica/Pharmacogenetics Research, Laboratorio SSL1, Centro Singular de Investigacio´n en Medicina Molecular y Enfermedades Cro´nicas (CiMUS), Santiago de Compostela, Spain. 4Department of Psychiatry, Faculty of Medicine, University of Toronto & Centre for Addiction & Mental Health (CAMH), Toronto, Canada. 5Programa Transtornos do Espectro Obsessivo-Compulsivo, Departamento e Instituto de Psiquiatria, Faculdade de Medicina, USP, Sa˜o Paulo, SP, Brazil. Objective: We investigated whether single nucleotide polymorphisms (SNPs) associated with neuroplasticity and activity of monoamine neurotransmitters, such as the brain-derived neurotrophic factor (BDNF, rs6265), the serotonin transporter (SLC6A4, rs25531), the tryptophan hydroxylase 1 (TPH1, rs1800532), the 5-hydroxytryptamine receptor 2A (HTR2A, rs6311, rs6313, rs7997012), and the catechol-O-methyltransferase (COMT, rs4680) genes, are associated with efficacy of transcranial direct current stimulation (tDCS) in major depression. -

Functions to Inhibit T Cell Activation BTNL2, a Butyrophilin-Like
BTNL2, a Butyrophilin-Like Molecule That Functions to Inhibit T Cell Activation Thang Nguyen, Xikui K. Liu, Yongliang Zhang and Chen Dong This information is current as of October 2, 2021. J Immunol 2006; 176:7354-7360; ; doi: 10.4049/jimmunol.176.12.7354 http://www.jimmunol.org/content/176/12/7354 Downloaded from References This article cites 40 articles, 10 of which you can access for free at: http://www.jimmunol.org/content/176/12/7354.full#ref-list-1 Why The JI? Submit online. http://www.jimmunol.org/ • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average by guest on October 2, 2021 Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2006 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology BTNL2, a Butyrophilin-Like Molecule That Functions to Inhibit T Cell Activation1 Thang Nguyen,2* Xikui K. Liu,2† Yongliang Zhang,† and Chen Dong3† B7 family members regulate T cell activation and tolerance. Although butyrophilin proteins share sequence homology with the B7 molecules, it is unclear whether they have any function in immune responses.