Free PDF Download
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Nuclear Organization and the Epigenetic Landscape of the Mus Musculus X-Chromosome Alicia Liu University of Connecticut - Storrs, [email protected]
University of Connecticut OpenCommons@UConn Doctoral Dissertations University of Connecticut Graduate School 8-9-2019 Nuclear Organization and the Epigenetic Landscape of the Mus musculus X-Chromosome Alicia Liu University of Connecticut - Storrs, [email protected] Follow this and additional works at: https://opencommons.uconn.edu/dissertations Recommended Citation Liu, Alicia, "Nuclear Organization and the Epigenetic Landscape of the Mus musculus X-Chromosome" (2019). Doctoral Dissertations. 2273. https://opencommons.uconn.edu/dissertations/2273 Nuclear Organization and the Epigenetic Landscape of the Mus musculus X-Chromosome Alicia J. Liu, Ph.D. University of Connecticut, 2019 ABSTRACT X-linked imprinted genes have been hypothesized to contribute parent-of-origin influences on social cognition. A cluster of imprinted genes Xlr3b, Xlr4b, and Xlr4c, implicated in cognitive defects, are maternally expressed and paternally silent in the murine brain. These genes defy classic mechanisms of autosomal imprinting, suggesting a novel method of imprinted gene regulation. Using Xlr3b and Xlr4c as bait, this study uses 4C-Seq on neonatal whole brain of a 39,XO mouse model, to provide the first in-depth analysis of chromatin dynamics surrounding an imprinted locus on the X-chromosome. Significant differences in long-range contacts exist be- tween XM and XP monosomic samples. In addition, XM interaction profiles contact a greater number of genes linked to cognitive impairment, abnormality of the nervous system, and abnormality of higher mental function. This is not a pattern that is unique to the imprinted Xlr3/4 locus. Additional Alicia J. Liu - University of Connecticut - 2019 4C-Seq experiments show that other genes on the X-chromosome, implicated in intellectual disability and/or ASD, also produce more maternal contacts to other X-linked genes linked to cognitive impairment. -

Modulated in Intestinal Inflammation A
BTNL2, a Butyrophilin/B7-Like Molecule, Is a Negative Costimulatory Molecule Modulated in Intestinal Inflammation This information is current as Heather A. Arnett, Sabine S. Escobar, Eva Gonzalez-Suarez, of September 28, 2021. Alison L. Budelsky, Lori A. Steffen, Norman Boiani, Ming Zhang, Gerald Siu, Avery W. Brewer and Joanne L. Viney J Immunol 2007; 178:1523-1533; ; doi: 10.4049/jimmunol.178.3.1523 http://www.jimmunol.org/content/178/3/1523 Downloaded from References This article cites 40 articles, 12 of which you can access for free at: http://www.jimmunol.org/content/178/3/1523.full#ref-list-1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication by guest on September 28, 2021 *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2007 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology BTNL2, a Butyrophilin/B7-Like Molecule, Is a Negative Costimulatory Molecule Modulated in Intestinal Inflammation Heather A. Arnett,1* Sabine S. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -
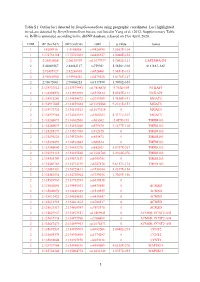
1 Table S1. Outlier Loci Detected by Deepgenomescan Using
Table S1. Outlier loci detected by DeepGenomeScan using geographic coordinates. Loci highlighted in red are detected by DeepGenomeScan but are not listed in Yang et al. (2012; Supplementary Table 4). RsID is annotated according to the dbSNP database released on 21st April, 2020. CHR BP (Grch37) BP (Grch38) rsID p.value Genes 1 1:4208918 1:4148858 rs9426495 3.8582E-104 1 1:175738168 1:175769032 rs6425357 2.0088E-105 2 2:20310668 2:20110907 rs11679737 1.2002E-111 LAPTM4A-DT 2 2:40289267 2:40062127 rs759361 5.5658E-104 SLC8A1-AS1 2 2:82495127 2:82268003 rs6726401 3.9451E-103 2 2:96660300 2:95994552 rs2579520 1.8176E-127 2 2:96672001 2:96006253 rs1917890 3.7698E-104 2 2:134333012 2:133575441 rs17816830 9.702E-105 NCKAP5 2 2:134350570 2:133592999 rs6715224 5.8745E-111 NCKAP5 2 2:134912243 2:134154672 rs2139309 1.7418E-191 MGAT5 2 2:134917005 2:134159434 rs11692586 9.2121E-151 MGAT5 2 2:134972732 2:134215161 rs11679218 0 MGAT5 2 2:134979966 2:134222395 rs1965183 5.3171E-107 MGAT5 2 2:135260071 2:134502500 rs503562 2.057E-153 TMEM163 2 2:135280039 2:134522468 rs579670 3.1477E-118 TMEM163 2 2:135285279 2:134527708 rs512375 0 TMEM163 2 2:135290221 2:134532650 rs655472 0 TMEM163 2 2:135290453 2:134532882 rs666614 0 TMEM163 2 2:135340840 2:134583270 rs842361 5.6717E-237 TMEM163 2 2:135393110 2:134635540 rs11684785 2.2164E-276 TMEM163 2 2:135430709 2:134673139 rs6745983 0 TMEM163 2 2:135469769 2:134712199 rs6747870 9.6117E-114 TMEM163 2 2:135483381 2:134725811 rs3739034 5.0357E-154 2 2:135483534 2:134725964 rs3739036 3.7269E-156 2 2:135539967 2:134782397 -

Genomic Signatures of Recent Adaptive Divergence in the Swamp Sparrow (Melospiza Georgiana)
GENOMIC SIGNATURES OF RECENT ADAPTIVE DIVERGENCE IN THE SWAMP SPARROW (MELOSPIZA GEORGIANA) A Dissertation Presented to the Faculty of the Graduate School of Cornell University In Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy by Petra Elizabeth Deane December 2017 © 2017 Petra Elizabeth Deane GENOMIC SIGNATURES OF RECENT ADAPTIVE DIVERGENCE IN THE SWAMP SPARROW (MELOSPIZA GEORGIANA) Petra Elizabeth Deane, Ph. D. Cornell University 2017 Populations that have recently diverged across sharp environmental gradients provide an opportunity to study the mechanisms by which natural selection drives adaptive divergence. Inland and coastal populations of the North American swamp sparrow (Melospiza georgiana) have become an emerging model system for studies of natural selection because they are morphologically and behaviorally distinct despite a very recent divergence time (<15,000 years), yet common garden experiments have demonstrated a genetic basis for their differences. I characterized genomic patterns of variation within and between inland and coastal swamp sparrows via reduced representation sequencing and demonstrated that background genomic differentiation (FST=0.02) and divergence (ΦST=0.05) between these populations is very low, rendering signatures of natural selection highly detectable (max FST=0.8). I then sequenced and assembled a de novo reference genome for the species and conducted a scan for genes involved in coastal adaptation, particularly the evolution of a deeper bill, darker plumage, and tolerance for salinity. I recovered a multigenic snapshot of adaptation via robust signatures of selection at 31 genes. As in Darwin’s finches, bone morphogenetic protein (BMP) signaling appears responsible for changes in bill depth, a putative magic trait for ecological speciation. -

Next Generation Exome Sequencing of Paediatric Inflammatory Bowel Disease Patients Identifies Rare and Novel Variants in Candida
Gut Online First, published on April 28, 2012 as 10.1136/gutjnl-2011-301833 Inflammatory bowel disease ORIGINAL ARTICLE Gut: first published as 10.1136/gutjnl-2011-301833 on 28 April 2012. Downloaded from Next generation exome sequencing of paediatric inflammatory bowel disease patients identifies rare and novel variants in candidate genes Katja Christodoulou,1 Anthony E Wiskin,2 Jane Gibson,1 William Tapper,1 Claire Willis,2 Nadeem A Afzal,3 Rosanna Upstill-Goddard,1 John W Holloway,4 Michael A Simpson,5 R Mark Beattie,3 Andrew Collins,1 Sarah Ennis1 < Additional materials are ABSTRACT published online only. To view Background Multiple genes have been implicated by Significance of this study these files please visit the association studies in altering inflammatory bowel journal online (http://gut.bmj. com/content/early/recent). disease (IBD) predisposition. Paediatric patients often What is already known on this subject? manifest more extensive disease and a particularly < For numbered affiliations see Genome-wide association studies have impli- end of article. severe disease course. It is likely that genetic cated numerous candidate genes for inflamma- predisposition plays a more substantial role in this group. tory bowel disease (IBD), but evidence of Correspondence to Objective To identify the spectrum of rare and novel causality for specific variants is largely absent. Dr Sarah Ennis, Genetic variation in known IBD susceptibility genes using exome Furthermore, by design, genome-wide associa- Epidemiology and Genomic sequencing analysis in eight individual cases of childhood Informatics Group, Human tion studies are limited to the study of Genetics, Faculty of Medicine, onset severe disease. -

Genome-Wide Analysis of DNA Methylation, Copy Number Variation, and Gene Expression in Monozygotic Twins Discordant for Primary Biliary Cirrhosis
UC Davis UC Davis Previously Published Works Title Genome-wide analysis of DNA methylation, copy number variation, and gene expression in monozygotic twins discordant for primary biliary cirrhosis. Permalink https://escholarship.org/uc/item/34d4m5nk Journal Frontiers in immunology, 5(MAR) ISSN 1664-3224 Authors Selmi, Carlo Cavaciocchi, Francesca Lleo, Ana et al. Publication Date 2014 DOI 10.3389/fimmu.2014.00128 Peer reviewed eScholarship.org Powered by the California Digital Library University of California ORIGINAL RESEARCH ARTICLE published: 28 March 2014 doi: 10.3389/fimmu.2014.00128 Genome-wide analysis of DNA methylation, copy number variation, and gene expression in monozygotic twins discordant for primary biliary cirrhosis Carlo Selmi 1,2*, Francesca Cavaciocchi 1,3, Ana Lleo4, Cristina Cheroni 5, Raffaele De Francesco5, Simone A. Lombardi 1, Maria De Santis 1,3, Francesca Meda1, Maria Gabriella Raimondo1, Chiara Crotti 1, Marco Folci 1, Luca Zammataro1, Marlyn J. Mayo6, Nancy Bach7, Shinji Shimoda8, Stuart C. Gordon9, Monica Miozzo10,11, Pietro Invernizzi 4, Mauro Podda1, Rossana Scavelli 5, Michelle R. Martin12, Michael F. Seldin13,14, Janine M. LaSalle 12 and M. Eric Gershwin2 1 Division of Rheumatology and Clinical Immunology, Humanitas Clinical and Research Center, Milan, Italy 2 Division of Rheumatology, Allergy, and Clinical Immunology, University of California at Davis, Davis, CA, USA 3 BIOMETRA Department, University of Milan, Milan, Italy 4 Liver Unit and Center for Autoimmune Liver Diseases, Humanitas Clinical and Research Center, Milan, Italy 5 National Institute of Molecular Genetics (INGM), Milan, Italy 6 University of Texas Southwestern, Dallas, TX, USA 7 Mt. Sinai University, NewYork, NY, USA 8 Clinical Research Center, National Nagasaki Medical Center, Nagasaki, Japan 9 Henry Ford Hospital, Detroit, MI, USA 10 Department of Pathophysiology and Transplantation, University of Milan, Milan, Italy 11 Division of Pathology, Fondazione IRCCS Cà Granda Ospedale Maggiore Policlinico, Milan, Italy 12 Genome Center and M.I.N.D. -

Familial Vs. Sporadic Sarcoidosis: BTNL2 Polymorphisms, Clinical
Familial vs. sporadic sarcoidosis: BTNL2 polymorphisms, clinical presentations, and outcomes in a French cohort Yves Pacheco, Alain Calender, Dominique Israël-Biet, Pascal Roy, Serge Lebecque, Vincent Cottin, Diane Bouvry, Hilario Nunes, Pascal Sève, Laurent Pérard, et al. To cite this version: Yves Pacheco, Alain Calender, Dominique Israël-Biet, Pascal Roy, Serge Lebecque, et al.. Familial vs. sporadic sarcoidosis: BTNL2 polymorphisms, clinical presentations, and outcomes in a French cohort. Orphanet Journal of Rare Diseases, BioMed Central, 2016, 11 (1), 10.1186/s13023-016-0546-4. hal- 01595465 HAL Id: hal-01595465 https://hal.archives-ouvertes.fr/hal-01595465 Submitted on 26 Sep 2017 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - ShareAlike| 4.0 International License Pacheco et al. Orphanet Journal of Rare Diseases (2016) 11:165 DOI 10.1186/s13023-016-0546-4 RESEARCH Open Access Familial vs. sporadic sarcoidosis: BTNL2 polymorphisms, clinical presentations, and outcomes in a French cohort Yves Pacheco1,11*, Alain Calender2, Dominique Israël-Biet3, Pascal Roy4, Serge Lebecque5, Vincent Cottin6, Diane Bouvry7, Hilario Nunes7, Pascal Sève8, Laurent Pérard9, Gilles Devouassoux8, Nathalie Freymond1, Chahira Khouatra6, Benoît Wallaert10, Raphaelle Lamy2, Mad-Hélénie Elsensohn4, Claire Bardel4, Dominique Valeyre7 and GSF group Abstract Background: The occurrence of familial forms of sarcoidosis (OMIM 181100) suggests a genetic predisposition. -

2027.Full.Pdf
Butyrophilin-like 2 Modulates B7 Costimulation To Induce Foxp3 Expression and Regulatory T Cell Development in Mature T Cells This information is current as of September 28, 2021. Ryan M. Swanson, Marc A. Gavin, Sabine S. Escobar, James B. Rottman, Brian P. Lipsky, Shishir Dube, Li Li, Jeannette Bigler, Martin Wolfson, Heather A. Arnett and Joanne L. Viney J Immunol 2013; 190:2027-2035; Prepublished online 28 Downloaded from January 2013; doi: 10.4049/jimmunol.1201760 http://www.jimmunol.org/content/190/5/2027 http://www.jimmunol.org/ Supplementary http://www.jimmunol.org/content/suppl/2013/01/28/jimmunol.120176 Material 0.DC1 References This article cites 40 articles, 18 of which you can access for free at: http://www.jimmunol.org/content/190/5/2027.full#ref-list-1 by guest on September 28, 2021 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. -

Mutation Induces Spontaneous Intestinal Inflammation in Mice
Yip1 domain family, member 6 (Yipf6) mutation induces spontaneous intestinal inflammation in mice Katharina Brandla, Wataru Tomisatoa,b, Xiaohong Lia,b, Christina Neppla, Elaine Piriea, Werner Falkc, Yu Xiaa, Eva Marie Y. Morescoa, Roberto Baccalad, Argyrios N. Theofilopoulosd, Bernd Schnable, and Bruce Beutlera,b,1 aDepartment of Genetics, The Scripps Research Institute, La Jolla, CA 92037; bCenter for Genetics of Host Defense, University of Texas Southwestern Medical Center, Dallas, TX 75390; cDepartment of Internal Medicine I, University of Regensburg, 93053 Regensburg, Germany; dDepartment of Immunology and Microbial Science, The Scripps Research Institute, La Jolla, CA 92037; and eDepartment of Medicine, University of California San Diego, La Jolla, CA 92093 Contributed by Bruce Beutler, June 19, 2012 (sent for review May 18, 2012) Using an environmentally sensitized genetic screen we identified affecting the structure of Mucin-2 also enhance susceptibility mutations that cause inflammatory colitis in mice. The X-linked to colitis, evidently by increasing ER stress (4), and do so in a Klein-Zschocher (KLZ) mutation created a null allele of Yipf6,a semidominant fashion. member of a gene family believed to regulate vesicular transport The fifth mutant, called Klein-Zschocher (KLZ) (after the in yeast, but without known functions in mammals. Yipf6 is a five town named in J. S. Bach cantata BWV 212), showed severe transmembrane-spanning protein associated with Golgi compart- weight loss after 1% DSS administration (Fig. 1B); both hemi- ments. Klein-Zschocher mutants were extremely sensitive to colitis zygous males and homozygous females displayed the phenotype induced by dextran sodium sulfate (DSS) and developed sponta- clearly. -

Functions to Inhibit T Cell Activation BTNL2, a Butyrophilin-Like
BTNL2, a Butyrophilin-Like Molecule That Functions to Inhibit T Cell Activation Thang Nguyen, Xikui K. Liu, Yongliang Zhang and Chen Dong This information is current as of October 2, 2021. J Immunol 2006; 176:7354-7360; ; doi: 10.4049/jimmunol.176.12.7354 http://www.jimmunol.org/content/176/12/7354 Downloaded from References This article cites 40 articles, 10 of which you can access for free at: http://www.jimmunol.org/content/176/12/7354.full#ref-list-1 Why The JI? Submit online. http://www.jimmunol.org/ • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average by guest on October 2, 2021 Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2006 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology BTNL2, a Butyrophilin-Like Molecule That Functions to Inhibit T Cell Activation1 Thang Nguyen,2* Xikui K. Liu,2† Yongliang Zhang,† and Chen Dong3† B7 family members regulate T cell activation and tolerance. Although butyrophilin proteins share sequence homology with the B7 molecules, it is unclear whether they have any function in immune responses.