Genome-Wide Analysis of DNA Methylation, Copy Number Variation, and Gene Expression in Monozygotic Twins Discordant for Primary Biliary Cirrhosis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cancer Immunity (1 December 2006) Vol
Cancer Immun 1424-9634Academy of Cancer Immunology Cancer Immunity (1 December 2006) Vol. 6, p. 12 Submitted: 26 September 2006. Accepted: 10 October 2006. Copyright © 2006 by Andrew J. G. Simpson 061012 Article Physical interaction of two cancer-testis antigens, MAGE-C1 (CT7) and NY-ESO-1 (CT6) Hearn J. Cho1*,**, Otavia L. Caballero2*, Sacha Gnjatic2, Valéria C. C. Andrade3, Gisele W. Colleoni3, Andre L. Vettore4, Hasina H. Outtz1, Sheila Fortunato2, Nasser Altorki1, Cathy A. Ferrera1, Ramon Chua2, Achim A. Jungbluth2, Yao-Tseng Chen1, Lloyd J. Old2 and Andrew J. G. Simpson2 1Weill Medical College of Cornell University, 1300 York Avenue, New York, NY 10021, USA 2Ludwig Institute for Cancer Research, New York Branch at Memorial Sloan-Kettering Cancer Center, 1275 York Avenue, New York, NY 10021, USA 3Escola Paulista de Medicina, Universidade Federal de Sao Paulo, Sao Paulo, SP, Brazil 4Ludwig Institute for Cancer Research, Sao Paulo Branch, Sao Paulo, SP, Brazil *These authors contributed equally to this work **Present address: NYU Cancer Institute, New York University School of Medicine, 550 First Avenue, New York, NY 10016, USA Contributed by: LJ Old Cancer/testis (CT) antigens are the protein products of germ line- encoded on the X chromosome (CT-X antigens) and those that associated genes that are activated in a wide variety of tumors and can are not (non-X CT antigens) (1). elicit autologous cellular and humoral immune responses. CT It is estimated that 10% of the genes on the X-chromosome antigens can be divided between those that are encoded on the X belong to CT-X families (5). -

Nuclear Organization and the Epigenetic Landscape of the Mus Musculus X-Chromosome Alicia Liu University of Connecticut - Storrs, [email protected]
University of Connecticut OpenCommons@UConn Doctoral Dissertations University of Connecticut Graduate School 8-9-2019 Nuclear Organization and the Epigenetic Landscape of the Mus musculus X-Chromosome Alicia Liu University of Connecticut - Storrs, [email protected] Follow this and additional works at: https://opencommons.uconn.edu/dissertations Recommended Citation Liu, Alicia, "Nuclear Organization and the Epigenetic Landscape of the Mus musculus X-Chromosome" (2019). Doctoral Dissertations. 2273. https://opencommons.uconn.edu/dissertations/2273 Nuclear Organization and the Epigenetic Landscape of the Mus musculus X-Chromosome Alicia J. Liu, Ph.D. University of Connecticut, 2019 ABSTRACT X-linked imprinted genes have been hypothesized to contribute parent-of-origin influences on social cognition. A cluster of imprinted genes Xlr3b, Xlr4b, and Xlr4c, implicated in cognitive defects, are maternally expressed and paternally silent in the murine brain. These genes defy classic mechanisms of autosomal imprinting, suggesting a novel method of imprinted gene regulation. Using Xlr3b and Xlr4c as bait, this study uses 4C-Seq on neonatal whole brain of a 39,XO mouse model, to provide the first in-depth analysis of chromatin dynamics surrounding an imprinted locus on the X-chromosome. Significant differences in long-range contacts exist be- tween XM and XP monosomic samples. In addition, XM interaction profiles contact a greater number of genes linked to cognitive impairment, abnormality of the nervous system, and abnormality of higher mental function. This is not a pattern that is unique to the imprinted Xlr3/4 locus. Additional Alicia J. Liu - University of Connecticut - 2019 4C-Seq experiments show that other genes on the X-chromosome, implicated in intellectual disability and/or ASD, also produce more maternal contacts to other X-linked genes linked to cognitive impairment. -
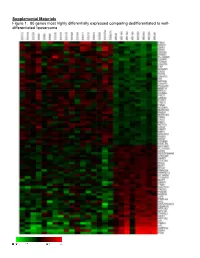
Supplemental Materials Figure 1. 80 Genes Most Highly Differentially Expressed Comparing Dedifferentiated to Well- Differentiated Liposarcoma
Supplemental Materials Figure 1. 80 genes most highly differentially expressed comparing dedifferentiated to well- differentiated liposarcoma Figure 2. Validation of microa CDC2, RACGAP1, FGFR-2, MAD2 and CITED1 200 CDK4 by Liposarcoma Subtype 16 0 12 0 80 Expression40 Level 0 Nl Fat rray data by quantitative RT-P Well Diff 16 0 p16 by Liposarcoma Subtype Dediff 12 0 80 Myxoid Expression40 Level Round Cell 0 12 0 MDM2 by Liposarcoma Subtype 10 0 Pleomorphic Nl Fat 80 60 40 Expression Level 20 RACGAP1 by LiposarcomaWell Diff Subtype 0 70 CR for genes CDK4, MDM2, p16, 60 Dediff 50 Nl Fat 40 30 Myxoid Expression20 Level Well Diff 10 50 0 Round Cell CDC2 by Liposarcoma Subtype Dediff 40 Nl Fat Pleomorphic 30 Myxoid 20 Expression Level Well Diff 10 MAD2L1 by Liposarcoma Subtype Round Cell 60 0 50 Dediff Pleomorphic 40 Nl Fat 30 Myxoid 20 Expression Level Well Diff 10 FGFR-2 by Liposarcoma Subtype 0 Round Cell 200 16 0 Dediff Nl Fat Pleomorphic 12 0 Myxoid 80 Expression Level Well Diff 40 Round Cell 0 Dediff Pleomorphic Nl Fat Myxoid Well Diff CITED1 by Liposarcoma Subtype Round Cell 10 0 80 Dediff Pleomorphic 60 40 Myxoid Expression Level 20 0 Round Cell Nl Fat Pleomorphic Well Diff Dediff Myxoid Round Cell Pleomorphic Table 1. 142 Gene classifier for liposarcoma subtype The genes used for each pair wise subtype comparison are grouped together. The flag column indicates which genes are unique to each subtype comparison. The values show the mean expression levels (actually the mean of the log expression levels was computed and than transformed back to absolute expression level). -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Produktinformation
Produktinformation Diagnostik & molekulare Diagnostik Laborgeräte & Service Zellkultur & Verbrauchsmaterial Forschungsprodukte & Biochemikalien Weitere Information auf den folgenden Seiten! See the following pages for more information! Lieferung & Zahlungsart Lieferung: frei Haus Bestellung auf Rechnung SZABO-SCANDIC Lieferung: € 10,- HandelsgmbH & Co KG Erstbestellung Vorauskassa Quellenstraße 110, A-1100 Wien T. +43(0)1 489 3961-0 Zuschläge F. +43(0)1 489 3961-7 [email protected] • Mindermengenzuschlag www.szabo-scandic.com • Trockeneiszuschlag • Gefahrgutzuschlag linkedin.com/company/szaboscandic • Expressversand facebook.com/szaboscandic SANTA CRUZ BIOTECHNOLOGY, INC. MAGE-D4/MAGE-D4B CRISPR/Cas9 KO Plasmid (h): sc-418394 BACKGROUND APPLICATIONS The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and MAGE-D4/MAGE-D4B CRISPR/Cas9 KO Plasmid (h) is recommended for the CRISPR-associated protein (Cas9) system is an adaptive immune response disruption of gene expression in human cells. defense mechanism used by archea and bacteria for the degradation of foreign genetic material (4,6). This mechanism can be repurposed for other 20 nt non-coding RNA sequence: guides Cas9 functions, including genomic engineering for mammalian systems, such as to a specific target location in the genomic DNA gene knockout (KO) (1,2,3,5). CRISPR/Cas9 KO Plasmid products enable the U6 promoter: drives gRNA scaffold: helps Cas9 identification and cleavage of specific genes by utilizing guide RNA (gRNA) expression of gRNA bind to target DNA sequences derived from the Genome-scale CRISPR Knock-Out (GeCKO) v2 library developed in the Zhang Laboratory at the Broad Institute (3,5). Termination signal Green Fluorescent Protein: to visually REFERENCES verify transfection CRISPR/Cas9 Knockout Plasmid CBh (chicken β-Actin 1. -

Expression of Cancer-Testis Antigens MAGEA1, MAGEA3, ACRBP, PRAME, SSX2, and CTAG2 in Myxoid and Round Cell Liposarcoma
Modern Pathology (2014) 27, 1238–1245 1238 & 2014 USCAP, Inc All rights reserved 0893-3952/14 $32.00 Expression of cancer-testis antigens MAGEA1, MAGEA3, ACRBP, PRAME, SSX2, and CTAG2 in myxoid and round cell liposarcoma Jessica A Hemminger1, Amanda Ewart Toland2, Thomas J Scharschmidt3, Joel L Mayerson3, Denis C Guttridge2 and O Hans Iwenofu1 1Department of Pathology and Laboratory Medicine, Wexner Medical Center at The Ohio State University, Columbus, OH, USA; 2Department of Molecular Virology, Immunology and Medical Genetics, The Ohio State University Wexner Medical Center, Columbus, OH, USA and 3Department of Orthopedics, The Ohio State University Wexner Medical Center, Columbus, OH, USA Myxoid and round-cell liposarcoma is a frequently encountered liposarcoma subtype. The mainstay of treatment remains surgical excision with or without chemoradiation. However, treatment options are limited in the setting of metastatic disease. Cancer-testis antigens are immunogenic antigens with the expression largely restricted to testicular germ cells and various malignancies, making them attractive targets for cancer immunotherapy. Gene expression studies have reported the expression of various cancer-testis antigens in liposarcoma, with mRNA expression of CTAG1B, CTAG2, MAGEA9, and PRAME described specifically in myxoid and round-cell liposarcoma. Herein, we further explore the expression of the cancer-testis antigens MAGEA1, ACRBP, PRAME, and SSX2 in myxoid and round-cell liposarcoma by immunohistochemistry in addition to determining mRNA levels of CTAG2 (LAGE-1), PRAME, and MAGEA3 by quantitative real-time PCR. Samples in formalin-fixed paraffin-embedded blocks (n ¼ 37) and frozen tissue (n ¼ 8) were obtained for immunohistochemistry and quantitative real-time PCR, respectively. Full sections were stained with antibodies to MAGEA1, ACRBP, PRAME, and SSX2 and staining was assessed for intensity (1–2 þ ) and percent tumor positivity. -

Secretion and LPS-Induced Endotoxin Shock Α Lipopolysaccharide
IFIT2 Is an Effector Protein of Type I IFN− Mediated Amplification of Lipopolysaccharide (LPS)-Induced TNF- α Secretion and LPS-Induced Endotoxin Shock This information is current as of September 27, 2021. Alexandra Siegfried, Susanne Berchtold, Birgit Manncke, Eva Deuschle, Julia Reber, Thomas Ott, Michaela Weber, Ulrich Kalinke, Markus J. Hofer, Bastian Hatesuer, Klaus Schughart, Valérie Gailus-Durner, Helmut Fuchs, Martin Hrabe de Angelis, Friedemann Weber, Mathias W. Hornef, Ingo B. Autenrieth and Erwin Bohn Downloaded from J Immunol published online 6 September 2013 http://www.jimmunol.org/content/early/2013/09/06/jimmun ol.1203305 http://www.jimmunol.org/ Supplementary http://www.jimmunol.org/content/suppl/2013/09/06/jimmunol.120330 Material 5.DC1 Why The JI? Submit online. by guest on September 27, 2021 • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Published September 6, 2013, doi:10.4049/jimmunol.1203305 The Journal of Immunology IFIT2 Is an Effector Protein of Type I IFN–Mediated Amplification of Lipopolysaccharide (LPS)-Induced TNF-a Secretion and LPS-Induced Endotoxin Shock Alexandra Siegfried,*,1 Susanne Berchtold,*,1 Birgit Manncke,* Eva Deuschle,* Julia Reber,* Thomas Ott,† Michaela Weber,‡ Ulrich Kalinke,x Markus J. -
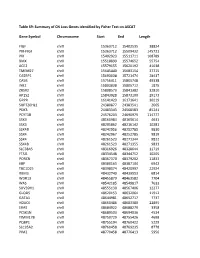
Table S9: Summary of CN Loss Genes Identified by Fisher Test on ASCAT
Table S9: Summary of CN Loss Genes identified by Fisher Test on ASCAT Gene Symbol Chromosome Start End Length FIGF chrX 15363712 15402535 38824 PIR-FIGF chrX 15363712 15509432 145721 PIR chrX 15402923 15511711 108789 BMX chrX 15518899 15574652 55754 ACE2 chrX 15579155 15620192 41038 TMEM27 chrX 15645440 15683154 37715 CA5BP1 chrX 15693038 15721474 28437 CA5B chrX 15756411 15805748 49338 INE2 chrX 15803838 15805712 1875 ZRSR2 chrX 15808573 15841382 32810 AP1S2 chrX 15843928 15873100 29173 GRPR chrX 16141423 16171641 30219 SUPT20HL1 chrX 24380877 24383541 2665 PDK3 chrX 24483343 24568583 85241 PCYT1B chrX 24576203 24690979 114777 SSX9 chrX 48160984 48165614 4631 SSX3 chrX 48205862 48216142 10281 SSX4B chrX 48242956 48252785 9830 SSX4 chrX 48242967 48252785 9819 SSX4 chrX 48261523 48271344 9822 SSX4B chrX 48261523 48271355 9833 SLC38A5 chrX 48316926 48328644 11719 FTSJ1 chrX 48334548 48344752 10205 PORCN chrX 48367370 48379202 11833 EBP chrX 48380163 48387104 6942 TBC1D25 chrX 48398074 48420997 22924 RBM3 chrX 48432740 48439553 6814 WDR13 chrX 48455879 48463582 7704 WAS chrX 48542185 48549817 7633 SUV39H1 chrX 48555130 48567406 12277 GLOD5 chrX 48620153 48632064 11912 GATA1 chrX 48644981 48652717 7737 HDAC6 chrX 48660486 48683380 22895 ERAS chrX 48684922 48688279 3358 PCSK1N chrX 48689503 48694036 4534 TIMM17B chrX 48750729 48755426 4698 PQBP1 chrX 48755194 48760422 5229 SLC35A2 chrX 48760458 48769235 8778 PIM2 chrX 48770458 48776413 5956 OTUD5 chrX 48779302 48815648 36347 KCND1 chrX 48818638 48828251 9614 GRIPAP1 chrX 48830133 48858675 28543 -

NY-ESO-1 (CTAG1B) Expression in Mesenchymal Tumors
Modern Pathology (2015) 28, 587–595 & 2015 USCAP, Inc. All rights reserved 0893-3952/15 $32.00 587 NY-ESO-1 (CTAG1B) expression in mesenchymal tumors Makoto Endo1,2,7, Marieke A de Graaff3,7, Davis R Ingram4, Simin Lim1, Dina C Lev4, Inge H Briaire-de Bruijn3, Neeta Somaiah5, Judith VMG Bove´e3, Alexander J Lazar6 and Torsten O Nielsen1 1Department of Pathology and Laboratory Medicine, University of British Columbia, Vancouver, British Columbia, Canada; 2Department of Orthopaedic Surgery, Kyushu University, Fukuoka, Japan; 3Department of Pathology, Leiden University Medical Center, Leiden, The Netherlands; 4Department of Surgical Oncology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA; 5Department of Sarcoma Medical Oncology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA and 6Department of Pathology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA New York esophageal squamous cell carcinoma 1 (NY-ESO-1, CTAG1B) is a cancer-testis antigen and currently a focus of several targeted immunotherapeutic strategies. We performed a large-scale immunohistochemical expression study of NY-ESO-1 using tissue microarrays of mesenchymal tumors from three institutions in an international collaboration. A total of 1132 intermediate and malignant and 175 benign mesenchymal lesions were enrolled in this study. Immunohistochemical staining was performed on tissue microarrays using a monoclonal antibody for NY-ESO-1. Among mesenchymal tumors, myxoid liposarcomas showed the highest positivity for NY-ESO-1 (88%), followed by synovial sarcomas (49%), myxofibrosarcomas (35%), and conventional chondrosarcomas (28%). Positivity of NY-ESO-1 in the remaining mesenchymal tumors was consistently low, and no immunoreactivity was observed in benign mesenchymal lesions. -

Small Cell Carcinoma of the Ovary, Hypercalcemic Type (SCCOHT) Beyond SMARCA4 Mutations: a Comprehensive Genomic Analysis
cells Article Small Cell Carcinoma of the Ovary, Hypercalcemic Type (SCCOHT) beyond SMARCA4 Mutations: A Comprehensive Genomic Analysis Aurélie Auguste 1,Félix Blanc-Durand 2 , Marc Deloger 3 , Audrey Le Formal 1, Rohan Bareja 4,5, David C. Wilkes 4 , Catherine Richon 6,Béatrice Brunn 2, Olivier Caron 6, Mojgan Devouassoux-Shisheboran 7,Sébastien Gouy 2, Philippe Morice 2, Enrica Bentivegna 2, Andrea Sboner 4,5,8, Olivier Elemento 4,8, Mark A. Rubin 9 , Patricia Pautier 2, Catherine Genestie 10, Joanna Cyrta 4,9,11 and Alexandra Leary 1,2,* 1 Medical Oncologist, Gynecology Unit, Lead Translational Research Team, INSERM U981, Gustave Roussy, 94805 Villejuif, France; [email protected] (A.A.); [email protected] (A.L.F.) 2 Gynecological Unit, Department of Medicine, Gustave Roussy, 94805 Villejuif, France; [email protected] (F.B.-D.); [email protected] (B.B.); [email protected] (S.G.); [email protected] (P.M.); [email protected] (E.B.); [email protected] (P.P.) 3 Bioinformatics Core Facility, Gustave Roussy Cancer Center, UMS CNRS 3655/INSERM 23 AMMICA, 94805 Villejuif, France; [email protected] 4 Caryl and Israel Englander Institute for Precision Medicine, Weill Cornell Medicine, New York, NY 10001, USA; [email protected] (R.B.); [email protected] (D.C.W.); [email protected] (A.S.); [email protected] (O.E.); [email protected] (J.C.) 5 Institute for Computational Biomedicine, Weill Cornell -

Entinostat Augments NK Cell Functions Via Epigenetic Upregulation of IFIT1-STING-STAT4 Pathway
www.oncotarget.com Oncotarget, 2020, Vol. 11, (No. 20), pp: 1799-1815 Research Paper Entinostat augments NK cell functions via epigenetic upregulation of IFIT1-STING-STAT4 pathway John M. Idso1, Shunhua Lao1, Nathan J. Schloemer1,2, Jeffrey Knipstein2, Robert Burns3, Monica S. Thakar1,2,* and Subramaniam Malarkannan1,2,4,5,* 1Laboratory of Molecular Immunology and Immunotherapy, Blood Research Institute, Versiti, Milwaukee, WI, USA 2Division of Pediatric Hematology-Oncology-BMT, Department of Pediatrics, Medical College of Wisconsin, Milwaukee, WI, USA 3Bioinformatics Core, Blood Research Institute, Versiti, Milwaukee, WI, USA 4Divson of Hematology-Oncology, Department of Medicine, Medical College of Wisconsin, Milwaukee, WI, USA 5Department of Microbiology and Immunology, Medical College of Wisconsin, Milwaukee, WI, USA *Co-senior authors Correspondence to: Monica S. Thakar, email: [email protected] Subramaniam Malarkannan, email: [email protected] Keywords: NK cells; histone deacetylase inhibitor; Ewing sarcoma; rhabdomyosarcoma; immunotherapy Received: September 10, 2019 Accepted: March 03, 2020 Published: May 19, 2020 Copyright: Idso et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Histone deacetylase inhibitors (HDACi) are an emerging cancer therapy; however, their effect on natural killer (NK) cell-mediated anti-tumor responses remain unknown. Here, we evaluated the impact of a benzamide HDACi, entinostat, on human primary NK cells as well as tumor cell lines. Entinostat significantly upregulated the expression of NKG2D, an essential NK cell activating receptor. Independently, entinostat augmented the expression of ULBP1, HLA, and MICA/B on both rhabdomyosarcoma and Ewing sarcoma cell lines. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase