Why the –Omic Future of Apicomplexa Should Include Gregarines Julie Boisard, Isabelle Florent
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Basal Body Structure and Composition in the Apicomplexans Toxoplasma and Plasmodium Maria E
Francia et al. Cilia (2016) 5:3 DOI 10.1186/s13630-016-0025-5 Cilia REVIEW Open Access Basal body structure and composition in the apicomplexans Toxoplasma and Plasmodium Maria E. Francia1* , Jean‑Francois Dubremetz2 and Naomi S. Morrissette3 Abstract The phylum Apicomplexa encompasses numerous important human and animal disease-causing parasites, includ‑ ing the Plasmodium species, and Toxoplasma gondii, causative agents of malaria and toxoplasmosis, respectively. Apicomplexans proliferate by asexual replication and can also undergo sexual recombination. Most life cycle stages of the parasite lack flagella; these structures only appear on male gametes. Although male gametes (microgametes) assemble a typical 9 2 axoneme, the structure of the templating basal body is poorly defined. Moreover, the rela‑ tionship between asexual+ stage centrioles and microgamete basal bodies remains unclear. While asexual stages of Plasmodium lack defined centriole structures, the asexual stages of Toxoplasma and closely related coccidian api‑ complexans contain centrioles that consist of nine singlet microtubules and a central tubule. There are relatively few ultra-structural images of Toxoplasma microgametes, which only develop in cat intestinal epithelium. Only a subset of these include sections through the basal body: to date, none have unambiguously captured organization of the basal body structure. Moreover, it is unclear whether this basal body is derived from pre-existing asexual stage centrioles or is synthesized de novo. Basal bodies in Plasmodium microgametes are thought to be synthesized de novo, and their assembly remains ill-defined. Apicomplexan genomes harbor genes encoding δ- and ε-tubulin homologs, potentially enabling these parasites to assemble a typical triplet basal body structure. -

Identification of a Novel Fused Gene Family Implicates Convergent
Chen et al. BMC Genomics (2018) 19:306 https://doi.org/10.1186/s12864-018-4685-y RESEARCH ARTICLE Open Access Identification of a novel fused gene family implicates convergent evolution in eukaryotic calcium signaling Fei Chen1,2,3, Liangsheng Zhang1, Zhenguo Lin4 and Zong-Ming Max Cheng2,3* Abstract Background: Both calcium signals and protein phosphorylation responses are universal signals in eukaryotic cell signaling. Currently three pathways have been characterized in different eukaryotes converting the Ca2+ signals to the protein phosphorylation responses. All these pathways have based mostly on studies in plants and animals. Results: Based on the exploration of genomes and transcriptomes from all the six eukaryotic supergroups, we report here in Metakinetoplastina protists a novel gene family. This family, with a proposed name SCAMK,comprisesSnRK3 fused calmodulin-like III kinase genes and was likely evolved through the insertion of a calmodulin-like3 gene into an SnRK3 gene by unequal crossover of homologous chromosomes in meiosis cell. Its origin dated back to the time intersection at least 450 million-year-ago when Excavata parasites, Vertebrata hosts, and Insecta vectors evolved. We also analyzed SCAMK’s unique expression pattern and structure, and proposed it as one of the leading calcium signal conversion pathways in Excavata parasite. These characters made SCAMK gene as a potential drug target for treating human African trypanosomiasis. Conclusions: This report identified a novel gene fusion and dated its precise fusion time -

The Planktonic Protist Interactome: Where Do We Stand After a Century of Research?
bioRxiv preprint doi: https://doi.org/10.1101/587352; this version posted May 2, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Bjorbækmo et al., 23.03.2019 – preprint copy - BioRxiv The planktonic protist interactome: where do we stand after a century of research? Marit F. Markussen Bjorbækmo1*, Andreas Evenstad1* and Line Lieblein Røsæg1*, Anders K. Krabberød1**, and Ramiro Logares2,1** 1 University of Oslo, Department of Biosciences, Section for Genetics and Evolutionary Biology (Evogene), Blindernv. 31, N- 0316 Oslo, Norway 2 Institut de Ciències del Mar (CSIC), Passeig Marítim de la Barceloneta, 37-49, ES-08003, Barcelona, Catalonia, Spain * The three authors contributed equally ** Corresponding authors: Ramiro Logares: Institute of Marine Sciences (ICM-CSIC), Passeig Marítim de la Barceloneta 37-49, 08003, Barcelona, Catalonia, Spain. Phone: 34-93-2309500; Fax: 34-93-2309555. [email protected] Anders K. Krabberød: University of Oslo, Department of Biosciences, Section for Genetics and Evolutionary Biology (Evogene), Blindernv. 31, N-0316 Oslo, Norway. Phone +47 22845986, Fax: +47 22854726. [email protected] Abstract Microbial interactions are crucial for Earth ecosystem function, yet our knowledge about them is limited and has so far mainly existed as scattered records. Here, we have surveyed the literature involving planktonic protist interactions and gathered the information in a manually curated Protist Interaction DAtabase (PIDA). In total, we have registered ~2,500 ecological interactions from ~500 publications, spanning the last 150 years. -

Hemosporidian Blood Parasites in Seabirds—A Comparative Genetic Study of Species from Antarctic to Tropical Habitats
Naturwissenschaften (2010) 97:809–817 DOI 10.1007/s00114-010-0698-3 ORIGINAL PAPER Hemosporidian blood parasites in seabirds—a comparative genetic study of species from Antarctic to tropical habitats Petra Quillfeldt & Javier Martínez & Janos Hennicke & Katrin Ludynia & Anja Gladbach & Juan F. Masello & Samuel Riou & Santiago Merino Received: 21 May 2010 /Revised: 7 July 2010 /Accepted: 7 July 2010 /Published online: 23 July 2010 # The Author(s) 2010. This article is published with open access at Springerlink.com Abstract Whereas some bird species are heavily affected by ranging from Antarctica to the tropical Indian Ocean. We did blood parasites in the wild, others reportedly are not. Seabirds, not detect parasites in 11 of these species, including one in particular, are often free from blood parasites, even in the Antarctic, four subantarctic, two temperate, and four tropical presence of potential vectors. By means of polymerase chain species. On the other hand, two subantarctic species, thin- reaction, we amplified a DNA fragment from the cytochrome billed prions Pachyptila belcheri and dolphin gulls Larus b gene to detect parasites of the genera Plasmodium, scoresbii, were found infected. One of 28 thin-billed prions Leucocytozoon,andHaemoproteus in 14 seabird species, had a Plasmodium infection whose DNA sequence was identical to lineage P22 of Plasmodium relictum, and one of 20 dolphin gulls was infected with a Haemoproteus lineage which appears phylogenetically clustered with parasites P. Quillfeldt (*) : K. Ludynia : A. Gladbach : J. F. Masello Max-Planck-Institut für Ornithologie, Vogelwarte Radolfzell, species isolated from passeriform birds such as Haemopro- Schlossallee 2, teus lanii, Haemoproteus magnus, Haemoproteus fringillae, 78315 Radolfzell, Germany Haemoproteus sylvae, Haemoproteus payevskyi,andHae- e-mail: [email protected] moproteus belopolskyi. -

University of Oklahoma
UNIVERSITY OF OKLAHOMA GRADUATE COLLEGE MACRONUTRIENTS SHAPE MICROBIAL COMMUNITIES, GENE EXPRESSION AND PROTEIN EVOLUTION A DISSERTATION SUBMITTED TO THE GRADUATE FACULTY in partial fulfillment of the requirements for the Degree of DOCTOR OF PHILOSOPHY By JOSHUA THOMAS COOPER Norman, Oklahoma 2017 MACRONUTRIENTS SHAPE MICROBIAL COMMUNITIES, GENE EXPRESSION AND PROTEIN EVOLUTION A DISSERTATION APPROVED FOR THE DEPARTMENT OF MICROBIOLOGY AND PLANT BIOLOGY BY ______________________________ Dr. Boris Wawrik, Chair ______________________________ Dr. J. Phil Gibson ______________________________ Dr. Anne K. Dunn ______________________________ Dr. John Paul Masly ______________________________ Dr. K. David Hambright ii © Copyright by JOSHUA THOMAS COOPER 2017 All Rights Reserved. iii Acknowledgments I would like to thank my two advisors Dr. Boris Wawrik and Dr. J. Phil Gibson for helping me become a better scientist and better educator. I would also like to thank my committee members Dr. Anne K. Dunn, Dr. K. David Hambright, and Dr. J.P. Masly for providing valuable inputs that lead me to carefully consider my research questions. I would also like to thank Dr. J.P. Masly for the opportunity to coauthor a book chapter on the speciation of diatoms. It is still such a privilege that you believed in me and my crazy diatom ideas to form a concise chapter in addition to learn your style of writing has been a benefit to my professional development. I’m also thankful for my first undergraduate research mentor, Dr. Miriam Steinitz-Kannan, now retired from Northern Kentucky University, who was the first to show the amazing wonders of pond scum. Who knew that studying diatoms and algae as an undergraduate would lead me all the way to a Ph.D. -

ABSTRACT Gregarine Parasitism in Dragonfly Populations of Central
ABSTRACT Gregarine Parasitism in Dragonfly Populations of Central Texas with an Assessment of Fitness Costs in Erythemis simplicicollis Jason L. Locklin, Ph.D. Mentor: Darrell S. Vodopich, Ph.D. Dragonfly parasites are widespread and frequently include gregarines (Phylum Apicomplexa) in the gut of the host. Gregarines are ubiquitous protozoan parasites that infect arthropods worldwide. More than 1,600 gregarine species have been described, but only a small percentage of invertebrates have been surveyed for these apicomplexan parasites. Some consider gregarines rather harmless, but recent studies suggest otherwise. Odonate-gregarine studies have more commonly involved damselflies, and some have considered gregarines to rarely infect dragonflies. In this study, dragonfly populations were surveyed for gregarines and an assessment of fitness costs was made in a common and widespread host species, Erythemis simplicicollis. Adult dragonfly populations were surveyed weekly at two reservoirs in close proximity to one another and at a flow-through wetland system. Gregarine prevalences and intensities were compared within host populations between genders, among locations, among wing loads, and through time. Host fitness parameters measured included wing load, egg size, clutch size, and total egg count. Of the 37 dragonfly species surveyed, 14 species (38%) hosted gregarines. Thirteen of those species were previously unreported as hosts. Gregarine prevalences ranged from 2% – 52%. Intensities ranged from 1 – 201. Parasites were aggregated among their hosts. Gregarines were found only in individuals exceeding a minimum wing load, indicating that gregarines are likely not transferred from the naiad to adult during emergence. Prevalence and intensity exhibited strong seasonality during both years at one of the reservoirs, but no seasonal trend was detected at the wetland. -

Black-Flies and Leucocytozoon Spp. As Causes of Mortality in Juvenile Great Horned Owls in the Yukon, Canada
Black-flies and Leucocytozoon spp. as Causes of Mortality in Juvenile Great Horned Owls in the Yukon, Canada D. Bruce Hunter1, Christoph Rohner2, and Doug C. Currie3 ABSTRACT.—Black fly feeding and infection with the blood parasite Leucocytozoon spp. caused mortality in juvenile Great Horned Owls (Bubo virginianus) in the Yukon, Canada during 1989-1990. The mortality occurred during a year of food shortage corresponding with the crash in snowshoe hare (Lepus americanus) populations. We postulate that the occurrence of disease was mediated by reduced food availability. Rohner (1994) evaluated the numerical re- black flies identified from Alaska, USA and the sponse of Great Horned Owls (Bubo virginianus) Yukon Territory, Canada, 36 percent are orni- to the snowshoe hare (Lepus americanus) cycle thophilic, 39 percent mammalophilic and 25 from 1988 to 1993 in the Kluane Lake area of percent autogenous (Currie 1997). Numerous southwestern Yukon, Canada. The survival of female black flies were obtained from the car- juvenile owls was very high during 1989 and casses of the juvenile owls, but only 45 of these 1990, both years of abundant hare populations. were sufficiently well preserved for identifica- Survival decreased in 1991, the first year of the tion. They belonged to four taxa as follows: snowshoe hare population decline (Rohner and Helodon (Distosimulium) pleuralis (Malloch), 1; Hunter 1996). Monitoring of nest sites Helodon (Parahelodon) decemarticulatus combined with tracking of individuals by radio- (Twinn), 3; Simulium (Eusimulium) aureum Fries telemetry provided us with carcasses of 28 ju- complex, 3; and Simulium (Eusimulium) venile owls found dead during 1990 and 1991 canonicolum (Dyar and Shannon) complex, 38 (Rohner and Doyle 1992). -
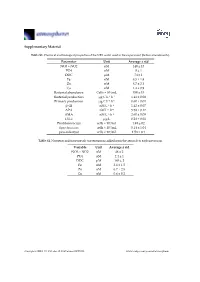
Supplementary Material Parameter Unit Average ± Std NO3 + NO2 Nm
Supplementary Material Table S1. Chemical and biological properties of the NRS water used in the experiment (before amendments). Parameter Unit Average ± std NO3 + NO2 nM 140 ± 13 PO4 nM 8 ± 1 DOC μM 74 ± 1 Fe nM 8.5 ± 1.8 Zn nM 8.7 ± 2.1 Cu nM 1.4 ± 0.9 Bacterial abundance Cells × 104/mL 350 ± 15 Bacterial production μg C L−1 h−1 1.41 ± 0.08 Primary production μg C L−1 h−1 0.60 ± 0.01 β-Gl nM L−1 h−1 1.42 ± 0.07 APA nM L−1 h−1 5.58 ± 0.17 AMA nM L−1·h−1 2.60 ± 0.09 Chl-a μg/L 0.28 ± 0.01 Prochlorococcus cells × 104/mL 1.49 ± 02 Synechococcus cells × 104/mL 5.14 ± 1.04 pico-eukaryot cells × 103/mL 1.58 × 0.1 Table S2. Nutrients and trace metals concentrations added from the aerosols to each mesocosm. Variable Unit Average ± std NO3 + NO2 nM 48 ± 2 PO4 nM 2.4 ± 1 DOC μM 165 ± 2 Fe nM 2.6 ± 1.5 Zn nM 6.7 ± 2.5 Cu nM 0.6 ± 0.2 Atmosphere 2019, 10, 358; doi:10.3390/atmos10070358 www.mdpi.com/journal/atmosphere Atmosphere 2019, 10, 358 2 of 6 Table S3. ANOVA test results between control, ‘UV-treated’ and ‘live-dust’ treatments at 20 h or 44 h, with significantly different values shown in bold. ANOVA df Sum Sq Mean Sq F Value p-value Chl-a 20 H 2, 6 0.03, 0.02 0.02, 0 4.52 0.0634 44 H 2, 6 0.02, 0 0.01, 0 23.13 0.002 Synechococcus Abundance 20 H 2, 7 8.23 × 107, 4.11 × 107 4.11 × 107, 4.51 × 107 0.91 0.4509 44 H 2, 7 5.31 × 108, 6.97 × 107 2.65 × 108, 1.16 × 107 22.84 0.0016 Prochlorococcus Abundance 20 H 2, 8 4.22 × 107, 2.11 × 107 2.11 × 107, 2.71 × 106 7.77 0.0216 44 H 2, 8 9.02 × 107, 1.47 × 107 4.51 × 107, 2.45 × 106 18.38 0.0028 Pico-eukaryote -

Supplementary Figure 1 Multicenter Randomised Control Trial 2746 Randomised
Supplementary Figure 1 Multicenter randomised control trial 2746 randomised 947 control 910 MNP without zinc 889 MNP with zinc 223 lost to follow up 219 lost to follow up 183 lost to follow up 34 refused 29 refused 37 refused 16 died 12 died 9 died 3 excluded 4 excluded 1 excluded 671 in follow-up 646 in follow-up 659 in follow-up at 24mo of age at 24mo of age at 24mo of age Selection for Microbiome sequencing 516 paired samples unavailable 469 paired samples unavailable 497 paired samples unavailable 69 antibiotic use 63 antibiotic use 67 antibiotic use 31 outside of WLZ criteria 37 outside of WLZ criteria 34 outside of WLZ criteria 6 diarrhea last 7 days 2 diarrhea last 7 days 7 diarrhea last 7 days 39 WLZ > -1 at 24 mo 10 WLZ < -2 at 24mo 58 WLZ > -1 at 24 mo 17 WLZ < -2 at 24mo 48 WLZ > -1 at 24 mo 8 WLZ < -2 at 24mo available for selection available for selection available for selection available for selection available for selection1 available for selection1 14 selected 10 selected 15 selected 14 selected 20 selected1 7 selected1 1 Two subjects (one in the reference WLZ group and one undernourished) had, at 12 months, no diarrhea within 1 day of stool collection but reported diarrhea within 7 days prior. Length, cm kg Weight, Supplementary Figure 2. Length (left) and weight (right) z-scores of children recruited into clinical trial NCT00705445 during the first 24 months of life. Median and quantile values are shown, with medians for participants profiled in current study indicated by red (undernourished) and black (reference WLZ) lines. -

Tracing the Origin of Planktonic Protists in an Ancient Lake
microorganisms Article Tracing the Origin of Planktonic Protists in an Ancient Lake Nataliia V. Annenkova 1,* , Caterina R. Giner 2,3 and Ramiro Logares 2,* 1 Limnological Institute Siberian Branch of the Russian Academy of Sciences 3, Ulan-Batorskaya St., 664033 Irkutsk, Russia 2 Institute of Marine Sciences (ICM), CSIC, Passeig Marítim de la Barceloneta, 37-49, ES08003 Barcelona, Spain; [email protected] 3 Institute for the Oceans and Fisheries, University of British Columbia, 2202 Main Mall, Vancouver, BC V6T 1Z4, Canada * Correspondence: [email protected] (N.V.A.); [email protected] (R.L.) Received: 26 February 2020; Accepted: 7 April 2020; Published: 9 April 2020 Abstract: Ancient lakes are among the most interesting models for evolution studies because their biodiversity is the result of a complex combination of migration and speciation. Here, we investigate the origin of single celled planktonic eukaryotes from the oldest lake in the world—Lake Baikal (Russia). By using 18S rDNA metabarcoding, we recovered 1414 Operational Taxonomic Units (OTUs) belonging to protists populating surface waters (1–50 m) and representing pico/nano-sized cells. The recovered communities resembled other lacustrine freshwater assemblages found elsewhere, especially the taxonomically unclassified protists. However, our results suggest that a fraction of Baikal protists could belong to glacial relicts and have close relationships with marine/brackish species. Moreover, our results suggest that rapid radiation may have occurred among some protist taxa, partially mirroring what was already shown for multicellular organisms in Lake Baikal. We found 16% of the OTUs belonging to potential species flocks in Stramenopiles, Alveolata, Opisthokonta, Archaeplastida, Rhizaria, and Hacrobia. -

Paraophioidina Scolecoides N. Sp., a New Aseptate Penaeus Vannamei
DISEASES OF AQUATIC ORGANISMS Vol. 19: 67-75,1994 Published June 9 Dis. aquat. Org. 1 l Paraophioidina scolecoides n. sp., a new aseptate gregarine from cultured Pacific white shrimp Penaeus vannamei Timothy C. Jonesl, Robin M. O~erstreet'~*,Jeffrey M. Lotzl, Paul F. Frelier2 'Gulf Coast Research Laboratory, PO Box 7000, Ocean Springs, Mississippi 39566, USA 2Department of Veterinary Pathobiology, College of Veterinary Medicine, Texas A&M University, College Station, Texas 77843, USA ABSTRACT: The aseptate gregarine Paraophloidina scolecoides n. sp. (Eugregarinorida: Lecud- inidae) heavily infected the nlidgut of cultured larval and postlarval specimens of Penaeus vannamei from a commercial 'seed-production' facility in Texas, USA. It is morphologically similar to P korot- neffiand P vibiliae, but it can be distinguished from them and from other members of the genus by having gamonts associated exclusively by lateral syzygy. Shrimp acquired the infection at the facility; nauph did not show any evidence of infection, but protozoea, mysis, and postlarval shrimp had a prevalence and intensity of infection ranging from 56 to 80 % and 10 to >50 parasites, respectively. Infected shrimp removed from the facility to aquaria at another location lost their gamont infection within 7 d. When voided from the gut, the gregarine disintegrated in seawater. Results suggest that P vannamei is an accidental host, although a survey of representative members of the invertebrate fauna from the environment associated with the facility failed to discover other hosts. No link was established between infection and either the broodstock or the water or detritus from the nursery or broodstock tanks. KEY WORDS: Gregarine . -

Studies in Cryptosporidium
STUDIES IN CRYPTOSPORIDIUM: MAINTENANCE OF STABLE POPULATIONS THROUGH IN VIVO PROPOGATION AND MOLECULAR DETECTION STRATEGIES DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of the Ohio State University By Norma E. Ramirez, M.P.H. ! ! ! ! The Ohio State University 2005 Dissertation Committee: Approved by Dr. Srinand Sreevatsan, Adviser Dr. Y.M. Saif ______________________________ Dr. Roger W. Stich Adviser Dr. Lucy A. Ward Graduate Program in Veterinary Preventive Medicine ABSTRACT Cryptosporidiosis, an infection caused by several genotypically and phenotypically diverse Cryptosporidium species, is a serious enteric disease of animals and humans worldwide. The current understanding of cryptosporidiosis, transmission, diagnosis, treatment and prevention measures for this disease is discussed. Contaminated water represents the major source of Cryptosporidium infections for humans. Manure from cattle can be a major source of Cryptosporidium oocysts. Oocysts transport to surface water can occur through direct fecal contamination, surface transport from land-applied manure or leaching through the soil to groundwater. Identification of Cryptosporidium species and genotypes facilitates determining the origin of the oocysts and to recognize sources of infection in outbreak situations and the risk factors associated with transmission. Very few studies have applied isolation methods to field samples because of difficulties with detection of oocysts in environmental samples. The objective of this study was to develop an easy method that can be applied to field samples to rapidly detect the presence of Cryptosporidium oocysts and identify their species. A molecular detection system that included an oocyst recovery method combined with spin column DNA extraction, followed by PCR- hybridization for detection and a Real-Time PCR-melting curve analysis for species ii assignment.