Supplementary Figure 1 Multicenter Randomised Control Trial 2746 Randomised
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Basal Body Structure and Composition in the Apicomplexans Toxoplasma and Plasmodium Maria E
Francia et al. Cilia (2016) 5:3 DOI 10.1186/s13630-016-0025-5 Cilia REVIEW Open Access Basal body structure and composition in the apicomplexans Toxoplasma and Plasmodium Maria E. Francia1* , Jean‑Francois Dubremetz2 and Naomi S. Morrissette3 Abstract The phylum Apicomplexa encompasses numerous important human and animal disease-causing parasites, includ‑ ing the Plasmodium species, and Toxoplasma gondii, causative agents of malaria and toxoplasmosis, respectively. Apicomplexans proliferate by asexual replication and can also undergo sexual recombination. Most life cycle stages of the parasite lack flagella; these structures only appear on male gametes. Although male gametes (microgametes) assemble a typical 9 2 axoneme, the structure of the templating basal body is poorly defined. Moreover, the rela‑ tionship between asexual+ stage centrioles and microgamete basal bodies remains unclear. While asexual stages of Plasmodium lack defined centriole structures, the asexual stages of Toxoplasma and closely related coccidian api‑ complexans contain centrioles that consist of nine singlet microtubules and a central tubule. There are relatively few ultra-structural images of Toxoplasma microgametes, which only develop in cat intestinal epithelium. Only a subset of these include sections through the basal body: to date, none have unambiguously captured organization of the basal body structure. Moreover, it is unclear whether this basal body is derived from pre-existing asexual stage centrioles or is synthesized de novo. Basal bodies in Plasmodium microgametes are thought to be synthesized de novo, and their assembly remains ill-defined. Apicomplexan genomes harbor genes encoding δ- and ε-tubulin homologs, potentially enabling these parasites to assemble a typical triplet basal body structure. -
Molecular Data and the Evolutionary History of Dinoflagellates by Juan Fernando Saldarriaga Echavarria Diplom, Ruprecht-Karls-Un
Molecular data and the evolutionary history of dinoflagellates by Juan Fernando Saldarriaga Echavarria Diplom, Ruprecht-Karls-Universitat Heidelberg, 1993 A THESIS SUBMITTED IN PARTIAL FULFILMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY in THE FACULTY OF GRADUATE STUDIES Department of Botany We accept this thesis as conforming to the required standard THE UNIVERSITY OF BRITISH COLUMBIA November 2003 © Juan Fernando Saldarriaga Echavarria, 2003 ABSTRACT New sequences of ribosomal and protein genes were combined with available morphological and paleontological data to produce a phylogenetic framework for dinoflagellates. The evolutionary history of some of the major morphological features of the group was then investigated in the light of that framework. Phylogenetic trees of dinoflagellates based on the small subunit ribosomal RNA gene (SSU) are generally poorly resolved but include many well- supported clades, and while combined analyses of SSU and LSU (large subunit ribosomal RNA) improve the support for several nodes, they are still generally unsatisfactory. Protein-gene based trees lack the degree of species representation necessary for meaningful in-group phylogenetic analyses, but do provide important insights to the phylogenetic position of dinoflagellates as a whole and on the identity of their close relatives. Molecular data agree with paleontology in suggesting an early evolutionary radiation of the group, but whereas paleontological data include only taxa with fossilizable cysts, the new data examined here establish that this radiation event included all dinokaryotic lineages, including athecate forms. Plastids were lost and replaced many times in dinoflagellates, a situation entirely unique for this group. Histones could well have been lost earlier in the lineage than previously assumed. -

The Planktonic Protist Interactome: Where Do We Stand After a Century of Research?
bioRxiv preprint doi: https://doi.org/10.1101/587352; this version posted May 2, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Bjorbækmo et al., 23.03.2019 – preprint copy - BioRxiv The planktonic protist interactome: where do we stand after a century of research? Marit F. Markussen Bjorbækmo1*, Andreas Evenstad1* and Line Lieblein Røsæg1*, Anders K. Krabberød1**, and Ramiro Logares2,1** 1 University of Oslo, Department of Biosciences, Section for Genetics and Evolutionary Biology (Evogene), Blindernv. 31, N- 0316 Oslo, Norway 2 Institut de Ciències del Mar (CSIC), Passeig Marítim de la Barceloneta, 37-49, ES-08003, Barcelona, Catalonia, Spain * The three authors contributed equally ** Corresponding authors: Ramiro Logares: Institute of Marine Sciences (ICM-CSIC), Passeig Marítim de la Barceloneta 37-49, 08003, Barcelona, Catalonia, Spain. Phone: 34-93-2309500; Fax: 34-93-2309555. [email protected] Anders K. Krabberød: University of Oslo, Department of Biosciences, Section for Genetics and Evolutionary Biology (Evogene), Blindernv. 31, N-0316 Oslo, Norway. Phone +47 22845986, Fax: +47 22854726. [email protected] Abstract Microbial interactions are crucial for Earth ecosystem function, yet our knowledge about them is limited and has so far mainly existed as scattered records. Here, we have surveyed the literature involving planktonic protist interactions and gathered the information in a manually curated Protist Interaction DAtabase (PIDA). In total, we have registered ~2,500 ecological interactions from ~500 publications, spanning the last 150 years. -

University of Oklahoma
UNIVERSITY OF OKLAHOMA GRADUATE COLLEGE MACRONUTRIENTS SHAPE MICROBIAL COMMUNITIES, GENE EXPRESSION AND PROTEIN EVOLUTION A DISSERTATION SUBMITTED TO THE GRADUATE FACULTY in partial fulfillment of the requirements for the Degree of DOCTOR OF PHILOSOPHY By JOSHUA THOMAS COOPER Norman, Oklahoma 2017 MACRONUTRIENTS SHAPE MICROBIAL COMMUNITIES, GENE EXPRESSION AND PROTEIN EVOLUTION A DISSERTATION APPROVED FOR THE DEPARTMENT OF MICROBIOLOGY AND PLANT BIOLOGY BY ______________________________ Dr. Boris Wawrik, Chair ______________________________ Dr. J. Phil Gibson ______________________________ Dr. Anne K. Dunn ______________________________ Dr. John Paul Masly ______________________________ Dr. K. David Hambright ii © Copyright by JOSHUA THOMAS COOPER 2017 All Rights Reserved. iii Acknowledgments I would like to thank my two advisors Dr. Boris Wawrik and Dr. J. Phil Gibson for helping me become a better scientist and better educator. I would also like to thank my committee members Dr. Anne K. Dunn, Dr. K. David Hambright, and Dr. J.P. Masly for providing valuable inputs that lead me to carefully consider my research questions. I would also like to thank Dr. J.P. Masly for the opportunity to coauthor a book chapter on the speciation of diatoms. It is still such a privilege that you believed in me and my crazy diatom ideas to form a concise chapter in addition to learn your style of writing has been a benefit to my professional development. I’m also thankful for my first undergraduate research mentor, Dr. Miriam Steinitz-Kannan, now retired from Northern Kentucky University, who was the first to show the amazing wonders of pond scum. Who knew that studying diatoms and algae as an undergraduate would lead me all the way to a Ph.D. -

ABSTRACT Gregarine Parasitism in Dragonfly Populations of Central
ABSTRACT Gregarine Parasitism in Dragonfly Populations of Central Texas with an Assessment of Fitness Costs in Erythemis simplicicollis Jason L. Locklin, Ph.D. Mentor: Darrell S. Vodopich, Ph.D. Dragonfly parasites are widespread and frequently include gregarines (Phylum Apicomplexa) in the gut of the host. Gregarines are ubiquitous protozoan parasites that infect arthropods worldwide. More than 1,600 gregarine species have been described, but only a small percentage of invertebrates have been surveyed for these apicomplexan parasites. Some consider gregarines rather harmless, but recent studies suggest otherwise. Odonate-gregarine studies have more commonly involved damselflies, and some have considered gregarines to rarely infect dragonflies. In this study, dragonfly populations were surveyed for gregarines and an assessment of fitness costs was made in a common and widespread host species, Erythemis simplicicollis. Adult dragonfly populations were surveyed weekly at two reservoirs in close proximity to one another and at a flow-through wetland system. Gregarine prevalences and intensities were compared within host populations between genders, among locations, among wing loads, and through time. Host fitness parameters measured included wing load, egg size, clutch size, and total egg count. Of the 37 dragonfly species surveyed, 14 species (38%) hosted gregarines. Thirteen of those species were previously unreported as hosts. Gregarine prevalences ranged from 2% – 52%. Intensities ranged from 1 – 201. Parasites were aggregated among their hosts. Gregarines were found only in individuals exceeding a minimum wing load, indicating that gregarines are likely not transferred from the naiad to adult during emergence. Prevalence and intensity exhibited strong seasonality during both years at one of the reservoirs, but no seasonal trend was detected at the wetland. -

Statistical Comparison of Excystation Methods in Cryptosporidium Parvum Oocysts
MASARYK UNIVERSITY FACULTY OF SCIENCE DEPT . OF BOTANY AND ZOOLOG Y BIOLOGICAL LY ACTIVE COMPOUNDS WIT H POTENTIAL ANTIPARASI TIC EFFECT AND THEIR IMPACT ON THE COURSE OF SELECTED PARASITOSES Ph.D. Dissertation Radka Pecková Supervisor: MVDr. Ivona Foitová, Ph.D . Brno 2018 Bibliographic Entry Author Mgr. Radka Pecková Faculty of Science, Masaryk University Department of Botany and Zoology Biological ly active compounds with potential Title of Thesis: antiparasitic effect and their impact on the processes of selected parasitoses Degree programme: Biology Field of Study: Parasitology Supervisor: MVDr. Ivona Foitová, Ph.D. Academic Year: 2017/2018 Number of Pages: 139 Keywords: Giardia intestinalis ; Cryptosporidium ; Anti - protozoal activity; Plant extracts; Drug of Choice; Natural Antiparasitics; Parasites; Archidendron fagifolium ; Diospyros sumatrana ; Piper betle ; Shorea sumatrana Bibliografický záznam Autor: Mgr. Radka Pecková Přírodovědecká fakulta, Masarykova univerzita Ústav botaniky a zoologie Biologicky aktivní látky s potencionálním Název práce: antiparazitárním účinkem a jejich působení na průběh vybraných parazitóz Studijní program: Biologie Studijní obor: Parazitologie Vedoucí práce: MVDr. Ivona Foitová, Ph.D. Akademický rok: 2017/2018 Počet stran: 139 Klíčová slova: Giardia intestinalis ; Cryptosporidium ; Antiprotozoární aktivita; Rostlinné extrakty; Alternativní léčiva; Přírodní antiparazitika; Paraziti; Archidendron fagifolium ; Diospyros sumatrana ; Piper betle ; Shorea sumatrana ABSTRAK T ABSTRACT This thesis deals with the study of the influence of extracts of selected plants from Indonesia on parasites Giardia intestinalis (Lambl) Alexeieff, 1914 and Cryptosporidium proliferans Kváč, Havrdová, Hlásková, Daňková, Kanděra, Ježková, Vítovec, Sak, Ortega, Xiao, Modrý, Jesudoss Chelladural, Prantlová & McEvoy, 2016 . Tested plants were selected based on behavioural data and the ability to reduce the intensity of parasitic infection in Sumatran orangutans. -

Metagenomic Characterization of Unicellular Eukaryotes in the Urban Thessaloniki Bay
Metagenomic characterization of unicellular eukaryotes in the urban Thessaloniki Bay George Tsipas SCHOOL OF ECONOMICS, BUSINESS ADMINISTRATION & LEGAL STUDIES A thesis submitted for the degree of Master of Science (MSc) in Bioeconomy Law, Regulation and Management May, 2019 Thessaloniki – Greece George Tsipas ’’Metagenomic characterization of unicellular eukaryotes in the urban Thessaloniki Bay’’ Student Name: George Tsipas SID: 268186037282 Supervisor: Prof. Dr. Savvas Genitsaris I hereby declare that the work submitted is mine and that where I have made use of another’s work, I have attributed the source(s) according to the Regulations set in the Student’s Handbook. May, 2019 Thessaloniki - Greece Page 2 of 63 George Tsipas ’’Metagenomic characterization of unicellular eukaryotes in the urban Thessaloniki Bay’’ 1. Abstract The present research investigates through metagenomics sequencing the unicellular protistan communities in Thermaikos Gulf. This research analyzes the diversity, composition and abundance in this marine environment. Water samples were collected monthly from April 2017 to February 2018 in the port of Thessaloniki (Harbor site, 40o 37’ 55 N, 22o 56’ 09 E). The extraction of DNA was completed as well as the sequencing was performed, before the downstream read processing and the taxonomic classification that was assigned using PR2 database. A total of 1248 Operational Taxonomic Units (OTUs) were detected but only 700 unicellular eukaryotes were analyzed, excluding unclassified OTUs, Metazoa and Streptophyta. In this research-based study the most abundant and diverse taxonomic groups were Dinoflagellata and Protalveolata. Specifically, the most abundant groups of all samples are Dinoflagellata with 190 OTUs (27.70%), Protalveolata with 139 OTUs (20.26%) Ochrophyta with 73 OTUs (10.64%), Cercozoa with 67 OTUs (9.77%) and Ciliophora with 64 OTUs (9.33%). -
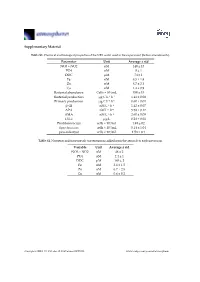
Supplementary Material Parameter Unit Average ± Std NO3 + NO2 Nm
Supplementary Material Table S1. Chemical and biological properties of the NRS water used in the experiment (before amendments). Parameter Unit Average ± std NO3 + NO2 nM 140 ± 13 PO4 nM 8 ± 1 DOC μM 74 ± 1 Fe nM 8.5 ± 1.8 Zn nM 8.7 ± 2.1 Cu nM 1.4 ± 0.9 Bacterial abundance Cells × 104/mL 350 ± 15 Bacterial production μg C L−1 h−1 1.41 ± 0.08 Primary production μg C L−1 h−1 0.60 ± 0.01 β-Gl nM L−1 h−1 1.42 ± 0.07 APA nM L−1 h−1 5.58 ± 0.17 AMA nM L−1·h−1 2.60 ± 0.09 Chl-a μg/L 0.28 ± 0.01 Prochlorococcus cells × 104/mL 1.49 ± 02 Synechococcus cells × 104/mL 5.14 ± 1.04 pico-eukaryot cells × 103/mL 1.58 × 0.1 Table S2. Nutrients and trace metals concentrations added from the aerosols to each mesocosm. Variable Unit Average ± std NO3 + NO2 nM 48 ± 2 PO4 nM 2.4 ± 1 DOC μM 165 ± 2 Fe nM 2.6 ± 1.5 Zn nM 6.7 ± 2.5 Cu nM 0.6 ± 0.2 Atmosphere 2019, 10, 358; doi:10.3390/atmos10070358 www.mdpi.com/journal/atmosphere Atmosphere 2019, 10, 358 2 of 6 Table S3. ANOVA test results between control, ‘UV-treated’ and ‘live-dust’ treatments at 20 h or 44 h, with significantly different values shown in bold. ANOVA df Sum Sq Mean Sq F Value p-value Chl-a 20 H 2, 6 0.03, 0.02 0.02, 0 4.52 0.0634 44 H 2, 6 0.02, 0 0.01, 0 23.13 0.002 Synechococcus Abundance 20 H 2, 7 8.23 × 107, 4.11 × 107 4.11 × 107, 4.51 × 107 0.91 0.4509 44 H 2, 7 5.31 × 108, 6.97 × 107 2.65 × 108, 1.16 × 107 22.84 0.0016 Prochlorococcus Abundance 20 H 2, 8 4.22 × 107, 2.11 × 107 2.11 × 107, 2.71 × 106 7.77 0.0216 44 H 2, 8 9.02 × 107, 1.47 × 107 4.51 × 107, 2.45 × 106 18.38 0.0028 Pico-eukaryote -

Predatory Flagellates – the New Recently Discovered Deep Branches of the Eukaryotic Tree and Their Evolutionary and Ecological Significance
Protistology 14 (1), 15–22 (2020) Protistology Predatory flagellates – the new recently discovered deep branches of the eukaryotic tree and their evolutionary and ecological significance Denis V. Tikhonenkov Papanin Institute for Biology of Inland Waters, Russian Academy of Sciences, Borok, 152742, Russia | Submitted March 20, 2020 | Accepted April 6, 2020 | Summary Predatory protists are poorly studied, although they are often representing important deep-branching evolutionary lineages and new eukaryotic supergroups. This short review/opinion paper is inspired by the recent discoveries of various predatory flagellates, which form sister groups of the giant eukaryotic clusters on phylogenetic trees, and illustrate an ancestral state of one or another supergroup of eukaryotes. Here we discuss their evolutionary and ecological relevance and show that the study of such protists may be essential in addressing previously puzzling evolutionary problems, such as the origin of multicellular animals, the plastid spread trajectory, origins of photosynthesis and parasitism, evolution of mitochondrial genomes. Key words: evolution of eukaryotes, heterotrophic flagellates, mitochondrial genome, origin of animals, photosynthesis, predatory protists, tree of life Predatory flagellates and diversity of eu- of the hidden diversity of protists (Moon-van der karyotes Staay et al., 2000; López-García et al., 2001; Edg- comb et al., 2002; Massana et al., 2004; Richards The well-studied multicellular animals, plants and Bass, 2005; Tarbe et al., 2011; de Vargas et al., and fungi immediately come to mind when we hear 2015). In particular, several prevailing and very abun- the term “eukaryotes”. However, these groups of dant ribogroups such as MALV, MAST, MAOP, organisms represent a minority in the real diversity MAFO (marine alveolates, stramenopiles, opistho- of evolutionary lineages of eukaryotes. -

Why the –Omic Future of Apicomplexa Should Include Gregarines Julie Boisard, Isabelle Florent
Why the –omic future of Apicomplexa should include Gregarines Julie Boisard, Isabelle Florent To cite this version: Julie Boisard, Isabelle Florent. Why the –omic future of Apicomplexa should include Gregarines. Biology of the Cell, Wiley, 2020, 10.1111/boc.202000006. hal-02553206 HAL Id: hal-02553206 https://hal.archives-ouvertes.fr/hal-02553206 Submitted on 24 Apr 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Article title: Why the –omic future of Apicomplexa should include Gregarines. Names of authors: Julie BOISARD1,2 and Isabelle FLORENT1 Authors affiliations: 1. Molécules de Communication et Adaptation des Microorganismes (MCAM, UMR 7245), Département Adaptations du Vivant (AVIV), Muséum National d’Histoire Naturelle, CNRS, CP52, 57 rue Cuvier 75231 Paris Cedex 05, France. 2. Structure et instabilité des génomes (STRING UMR 7196 CNRS / INSERM U1154), Département Adaptations du vivant (AVIV), Muséum National d'Histoire Naturelle, CP 26, 57 rue Cuvier 75231 Paris Cedex 05, France. Short Title: Gregarines –omics for Apicomplexa studies -

Tracing the Origin of Planktonic Protists in an Ancient Lake
microorganisms Article Tracing the Origin of Planktonic Protists in an Ancient Lake Nataliia V. Annenkova 1,* , Caterina R. Giner 2,3 and Ramiro Logares 2,* 1 Limnological Institute Siberian Branch of the Russian Academy of Sciences 3, Ulan-Batorskaya St., 664033 Irkutsk, Russia 2 Institute of Marine Sciences (ICM), CSIC, Passeig Marítim de la Barceloneta, 37-49, ES08003 Barcelona, Spain; [email protected] 3 Institute for the Oceans and Fisheries, University of British Columbia, 2202 Main Mall, Vancouver, BC V6T 1Z4, Canada * Correspondence: [email protected] (N.V.A.); [email protected] (R.L.) Received: 26 February 2020; Accepted: 7 April 2020; Published: 9 April 2020 Abstract: Ancient lakes are among the most interesting models for evolution studies because their biodiversity is the result of a complex combination of migration and speciation. Here, we investigate the origin of single celled planktonic eukaryotes from the oldest lake in the world—Lake Baikal (Russia). By using 18S rDNA metabarcoding, we recovered 1414 Operational Taxonomic Units (OTUs) belonging to protists populating surface waters (1–50 m) and representing pico/nano-sized cells. The recovered communities resembled other lacustrine freshwater assemblages found elsewhere, especially the taxonomically unclassified protists. However, our results suggest that a fraction of Baikal protists could belong to glacial relicts and have close relationships with marine/brackish species. Moreover, our results suggest that rapid radiation may have occurred among some protist taxa, partially mirroring what was already shown for multicellular organisms in Lake Baikal. We found 16% of the OTUs belonging to potential species flocks in Stramenopiles, Alveolata, Opisthokonta, Archaeplastida, Rhizaria, and Hacrobia. -

Assessing the Efficiency of Molecular Markers for the Species Identification of Gregarines Isolated from the Mealworm and Super Worm Midgut
Microorganisms 2018, 6, 119 1 of 20 Article Assessing the Efficiency of Molecular Markers for the Species Identification of Gregarines Isolated from the Mealworm and Super Worm Midgut Chiara Nocciolini 1, Claudio Cucini 2, Chiara Leo 2, Valeria Francardi 3, Elena Dreassi 4 and Antonio Carapelli 2,* 1 Polo d’Innovazione di Genomica Genetica e Biologia, 53100 Siena, Italy; [email protected] 2 Department of Life Sciences, University of Siena, 53100 Siena, Italy; [email protected] (C.C.); [email protected] (C.L.) 3 Consiglio per la ricerca in agricoltura e analisi dell’economia agraria – Centro di Difesa e Certificazione CREA-DC) Research Centre for Plant Protection and Certification, via di Lanciola 12/A, 50125 Firenze, Italy; [email protected] 4 Department of Biotechnology, Chemistry and Pharmacy, University of Siena, 53100 Siena, Italy; [email protected] * Correspondence: [email protected]; Tel.: +39-0577-234-410 Received: 28 September 2018; Accepted: 23 November 2018; Published: 27 November 2018 Abstract: Protozoa, of the taxon Gregarinasina, are a heterogeneous group of Apicomplexa that includes ~1600 species. They are parasites of a large variety of both marine and terrestrial invertebrates, mainly annelids, arthropods and mollusks. Unlike coccidians and heamosporidians, gregarines have not proven to have a negative effect on human welfare; thus, they have been poorly investigated. This study focuses on the molecular identification and phylogeny of the gregarine species found in the midgut of two insect species that are considered as an alternative source of animal proteins for the human diet: the mealworm Tenebrio molitor, and the super-worm Zophobas atratus (Coleoptera: Tenebrionidae).