Effect of Disease State on Human Carboxylesterase 1 Expression and Activity
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Carboxylesterase 1 / CES1 Antibody (Internal) Goat Polyclonal Antibody Catalog # ALS12250
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 Carboxylesterase 1 / CES1 Antibody (Internal) Goat Polyclonal Antibody Catalog # ALS12250 Specification Carboxylesterase 1 / CES1 Antibody (Internal) - Product Information Application WB, IHC Primary Accession P23141 Reactivity Human Host Goat Clonality Polyclonal Calculated MW 63kDa KDa Carboxylesterase 1 / CES1 Antibody (Internal) - Additional Information Gene ID 1066 Antibody (0.03 ug/ml) staining of Human Liver lysate (35 ug protein in RIPA buffer). Other Names Liver carboxylesterase 1, Acyl-coenzyme A:cholesterol acyltransferase, ACAT, Brain carboxylesterase hBr1, Carboxylesterase 1, CE-1, hCE-1, 3.1.1.1, Cocaine carboxylesterase, Egasyn, HMSE, Methylumbelliferyl-acetate deacetylase 1, 3.1.1.56, Monocyte/macrophage serine esterase, Retinyl ester hydrolase, REH, Serine esterase 1, Triacylglycerol hydrolase, TGH, CES1, CES2, SES1 Target/Specificity Human CES1. This antibody is expected to recognise all three reported isoforms (NP_001020366.1; NP_001020365.1; Anti-CES1 antibody IHC of human lung. NP_001257.4). Reconstitution & Storage Carboxylesterase 1 / CES1 Antibody Store at -20°C. Minimize freezing and (Internal) - Background thawing. Involved in the detoxification of xenobiotics Precautions and in the activation of ester and amide Carboxylesterase 1 / CES1 Antibody prodrugs. Hydrolyzes aromatic and aliphatic (Internal) is for research use only and not esters, but has no catalytic activity toward for use in diagnostic or therapeutic amides or a fatty acyl-CoA ester. Hydrolyzes procedures. the methyl ester group of cocaine to form benzoylecgonine. Catalyzes the transesterification of cocaine to form Carboxylesterase 1 / CES1 Antibody (Internal) - cocaethylene. Displays fatty acid ethyl ester Protein Information synthase activity, catalyzing the ethyl esterification of oleic acid to ethyloleate. -

Current Drug Metabolism, 2019, 20, 91-102
Send Orders for Reprints to [email protected] 91 Current Drug Metabolism, 2019, 20, 91-102 REVIEW ARTICLE ISSN: 1389-2002 eISSN: 1875-5453 Current Drug Impact Factor: Metabolism 2.655 The Impact of Carboxylesterases in Drug Metabolism and Pharmacokinetics The international journal for timely in-depth reviews on Drug Metabolism BENTHAM SCIENCE Li Di* Pfizer Inc., Eastern Point Road, Groton, Connecticut, CT 06354, USA Abstract: Background: Carboxylesterases (CES) play a critical role in catalyzing hydrolysis of esters, amides, car- bamates and thioesters, as well as bioconverting prodrugs and soft drugs. The unique tissue distribution of CES en- zymes provides great opportunities to design prodrugs or soft drugs for tissue targeting. Marked species differences in CES tissue distribution and catalytic activity are particularly challenging in human translation. Methods: Review and summarization of CES fundamentals and applications in drug discovery and development. A R T I C L E H I S T O R Y Results: Human CES1 is one of the most highly expressed drug metabolizing enzymes in the liver, while human intestine only expresses CES2. CES enzymes have moderate to high inter-individual variability and exhibit low to no expression in the fetus, but increase substantially during the first few months of life. The CES genes are highly po- Received: June 04, 2018 Revised: August 03, 2018 lymorphic and some CES genetic variants show significant influence on metabolism and clinical outcome of certain Accepted: August 08, 2018 drugs. Monkeys appear to be more predictive of human pharmacokinetics for CES substrates than other species. Low risk of clinical drug-drug interaction is anticipated for CES, although they should not be overlooked, particularly DOI: 10.2174/1389200219666180821094502 interaction with alcohols. -

The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease Jonathan Z
REVIEW pubs.acs.org/CR The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease Jonathan Z. Long* and Benjamin F. Cravatt* The Skaggs Institute for Chemical Biology and Department of Chemical Physiology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, California 92037, United States CONTENTS 2.4. Other Phospholipases 6034 1. Introduction 6023 2.4.1. LIPG (Endothelial Lipase) 6034 2. Small-Molecule Hydrolases 6023 2.4.2. PLA1A (Phosphatidylserine-Specific 2.1. Intracellular Neutral Lipases 6023 PLA1) 6035 2.1.1. LIPE (Hormone-Sensitive Lipase) 6024 2.4.3. LIPH and LIPI (Phosphatidic Acid-Specific 2.1.2. PNPLA2 (Adipose Triglyceride Lipase) 6024 PLA1R and β) 6035 2.1.3. MGLL (Monoacylglycerol Lipase) 6025 2.4.4. PLB1 (Phospholipase B) 6035 2.1.4. DAGLA and DAGLB (Diacylglycerol Lipase 2.4.5. DDHD1 and DDHD2 (DDHD Domain R and β) 6026 Containing 1 and 2) 6035 2.1.5. CES3 (Carboxylesterase 3) 6026 2.4.6. ABHD4 (Alpha/Beta Hydrolase Domain 2.1.6. AADACL1 (Arylacetamide Deacetylase-like 1) 6026 Containing 4) 6036 2.1.7. ABHD6 (Alpha/Beta Hydrolase Domain 2.5. Small-Molecule Amidases 6036 Containing 6) 6027 2.5.1. FAAH and FAAH2 (Fatty Acid Amide 2.1.8. ABHD12 (Alpha/Beta Hydrolase Domain Hydrolase and FAAH2) 6036 Containing 12) 6027 2.5.2. AFMID (Arylformamidase) 6037 2.2. Extracellular Neutral Lipases 6027 2.6. Acyl-CoA Hydrolases 6037 2.2.1. PNLIP (Pancreatic Lipase) 6028 2.6.1. FASN (Fatty Acid Synthase) 6037 2.2.2. PNLIPRP1 and PNLIPR2 (Pancreatic 2.6.2. -

Isolation and Characterization of the Prolyl Aminopeptidase Gene (Pap) from Aeromonas Sobria: Comparison with the Bacillus Coagulans Enzyme1
J. Biochem. 116, 818-825 (1994) Isolation and Characterization of the Prolyl Aminopeptidase Gene (pap) from Aeromonas sobria: Comparison with the Bacillus coagulans Enzyme1 Ana Kitazono,* Atsuko Kitano,* Daisuke Tsuru,•õ and Tadashi Yoshimoto*,2 *School of Pharmaceutical Sciences , Nagasaki University, 1-14 Bunkyo-machi, Nagasaki, Nagasaki 852; and •õ Department of Applied Microbiology, Kumamoto Institute of Technology, 4-22-1 Ikeda, Kumamoto, Kumamoto 860 Received for publication, May 16, 1994 The Aeromonas sobria pap gene encoding prolyl aminopeptidase (PAP) was cloned. It consists of 425 codons and encodes a homotetrameric enzyme of 205kDa. The purified enzyme showed an almost absolute specificity for amino-terminal proline. Proline and hydroxyproline residues from many peptide and amide substrates could be easily removed, while no activity was detected for substrates having other amino terminals. The enzyme was very similar to that from Bacillus coagulans in many aspects, such as the strong inhibition caused by PCMB and the weak or no inhibition caused by DFP and chelators, respectively. However, these enzymes show only 15% identity in their amino acid sequences. Differences were also observed in their molecular weight, stability and activity toward some peptide substrates. When aligning the deduced amino acid sequence with known sequences from other microorganisms, conserved sequences were found at the amino-terminal region; the significance of these conserved regions is discussed. Based on the results of this work, and on the studies available to date, the occurrence of at least two types of PAPs is postulated. One group would be formed by the Bacillus, Neisseria, and Lactobacillus enzymes, and the other by enzymes such as the Aeromonas PAP. -

P-Glycoprotein, CYP3A, and Plasma Carboxylesterase Determine Brain and Blood Disposition of the Mtor Inhibitor Everolimus (Afinitor) in Mice
Published OnlineFirst April 11, 2014; DOI: 10.1158/1078-0432.CCR-13-1759 Clinical Cancer Cancer Therapy: Preclinical Research P-Glycoprotein, CYP3A, and Plasma Carboxylesterase Determine Brain and Blood Disposition of the mTOR Inhibitor Everolimus (Afinitor) in Mice Seng Chuan Tang1, Rolf W. Sparidans3, Ka Lei Cheung4, Tatsuki Fukami5, Selvi Durmus1, Els Wagenaar1, Tsuyoshi Yokoi5, Bart J.M. van Vlijmen4, Jos H. Beijnen2,3, and Alfred H. Schinkel1 Abstract Purpose: To clarify the role of ABCB1, ABCG2, and CYP3A in blood and brain exposure of everolimus using knockout mouse models. À À À À À À À À Experimental Design: We used wild-type, Abcb1a/1b / , Abcg2 / , Abcb1a/1b;Abcg2 / , and Cyp3a / mice to study everolimus oral bioavailability and brain accumulation. Results: Following everolimus administration, brain concentrations and brain-to-liver ratios were À À À À À À substantially increased in Abcb1a/1b / and Abcb1a/1b;Abcg2 / , but not Abcg2 / mice. The fraction of everolimus located in the plasma compartment was highly increased in all knockout strains. In vitro, everolimus was rapidly degraded in wild-type but not knockout plasma. Carboxylesterase 1c (Ces1c), a plasma carboxylesterase gene, was highly upregulated (80-fold) in the liver of knockout mice relative to wild-type mice, and plasma Ces1c likely protected everolimus from degradation by binding and stabilizing it. This binding was prevented by preincubation with the carboxylesterase inhibitor BNPP. In vivo knockdown experiments confirmed the involvement of Ces1c in everolimus stabilization. Everolimus also markedly inhibited the hydrolysis of irinotecan and p-nitrophenyl acetate by mouse plasma carboxylesterase À À and recombinant human CES2, respectively. -

Baboon JMP CES Paper
Baboon carboxylesterases 1 and 2: sequences, structures and phylogenetic relationships with human and other primate carboxylesterases Author Holmes, Roger S, Glenn, Jeremy P, VandeBerg, John L, Cox, Laura A Published 2009 Journal Title Journal of Medical Primatology DOI https://doi.org/10.1111/j.1600-0684.2008.00315.x Copyright Statement © 2009 John Wiley & Sons A/S. This is the author-manuscript version of the paper. Reproduced in accordance with the copyright policy of the publisher.The definitive version is available at www.interscience.wiley.com Downloaded from http://hdl.handle.net/10072/29362 Griffith Research Online https://research-repository.griffith.edu.au Baboon Carboxylesterases 1 and 2: Sequences, Structures and Phylogenetic Relationships with Human and other Primate Carboxylesterases Roger S. Holmes 1,2,3 , Jeremy P. Glenn 1, John L. VandeBerg 1,2 , Laura A. Cox 1,2,4 1. Department of Genetics and 2. Southwest National Primate Research Center, Southwest Foundation for Biomedical Research, San Antonio, TX, USA, and 3. School of Biomolecular and Physical Sciences, Griffith University, Nathan. Queensland, Australia 4. Corresponding Author: Laura A. Cox, Ph.D. Department of Genetics Southwest National Primate Research Center Southwest Foundation for Biomedical Research San Antonio, TX, USA 78227 Email: [email protected] Phone: 210-258-9687 Fax: 210-258-9600 Keywords: cDNA sequence; amino acid sequence; 3-D structure Running Head: Carboxylesterases: sequences and phylogeny Published in Journal of Medical Primatology (2009) 38: 27-38. Abstract Background Carboxylesterase (CES) is predominantly responsible for the detoxification of a wide range of drugs and narcotics, and catalyze several reactions in cholesterol and fatty acid metabolism. -
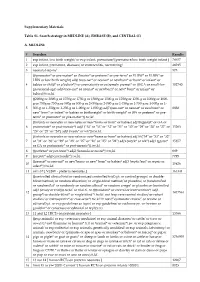
EMBASE (B), and CENTRAL (C) A. MEDLINE # Searches
Supplementary Materials Table S1. Search strategy in MEDLINE (A), EMBASE (B), and CENTRAL (C) A. MEDLINE # Searches Results 1 exp infant, low birth weight/ or exp infant, premature/ [premature/low birth weight infant ] 78657 2 exp infant, premature, diseases/ or enterocolitis, necrotizing/ 46015 3 neonatal sepsis/ 575 (((prematur* or pre-matur* or i?matur* or preterm* or pre-term* or VLBW* or ELBW* or LBW or low birth weight) adj6 (neo-nat* or neonat* or newborn* or born* or infant* or 4 babies or child* or p?ediatr*)) or prematurity or extremely premat* or ((SGA or small-for- 102740 gestational-age) adj6 (neo-nat* or neonat* or newborn* or new* born* or infant* or babies))).tw,kf. ((2000g or 2000-g or 1750g or 1750-g or 1500g or 1500-g or 1250g or 1250-g or 1000g or 1000- g or 750g or 750-g or 500g or 500-g or 2-000g or 2-000-g or 1-750g or 1-750-g or 1-500g or 1- 5 500-g or 1-250g or 1-250-g or 1-000g or 1-000-g) adj7 (neo-nat* or neonat* or newborn* or 8838 new* born* or infant* or babies or birthweight* or birth weight* or BW or preterm* or pre- term* or prematur* or pre-matur*)).tw,kf. ((infants or neonates or neo-nates or new*borns or born* or babies) adj18 (gestat* or GA or 6 postmenstr* or post-menstr*) adj3 ("34" or "33" or "32" or "31" or "30" or "29" or "28" or "27" or 15263 "26" or "25" or "24") adj3 (week* or wk*)).tw,kf. -

The Role of the B-Type Phospholipases in S. Cerevisiae: Function, Regulation, and Physiological Relevance in Lipid Homeostasis Beth a Surlow
Duquesne University Duquesne Scholarship Collection Electronic Theses and Dissertations Summer 2014 The Role of the B-Type Phospholipases in S. cerevisiae: Function, Regulation, and Physiological Relevance in Lipid Homeostasis Beth A Surlow Follow this and additional works at: https://dsc.duq.edu/etd Recommended Citation Surlow, B. (2014). The Role of the B-Type Phospholipases in S. cerevisiae: Function, Regulation, and Physiological Relevance in Lipid Homeostasis (Doctoral dissertation, Duquesne University). Retrieved from https://dsc.duq.edu/etd/1255 This Immediate Access is brought to you for free and open access by Duquesne Scholarship Collection. It has been accepted for inclusion in Electronic Theses and Dissertations by an authorized administrator of Duquesne Scholarship Collection. For more information, please contact [email protected]. THE ROLE OF B-TYPE PHOSPHOLIPASES IN S. CEREVISIAE: FUNCTION, REGULATION, AND PHYSIOLOGICAL RELEVANCE IN LIPID HOMEOSTASIS A Dissertation Submitted to the Bayer School of Natural and Environmental Sciences Duquesne University In partial fulfillment of the requirements for the degree of Doctor of Philosophy By Beth A. Surlow August 2014 Copyright by Beth A. Surlow 2014 THE ROLE OF B-TYPE PHOSPHOLIPASES IN S. CEREVISIAE: FUNCTION, REGULATION, AND PHYSIOLOGICAL RELEVANCE IN LIPID HOMEOSTASIS By Beth A. Surlow Approved June 5, 2014 ________________________________ ________________________________ Dr. Jana Patton-Vogt Dr. Philip Auron Associate Professor of Biological Professor of Biological Sciences Sciences (Committee Member) (Committee Chair) ________________________________ ________________________________ Dr. Joseph McCormick Dr. Jeffrey Brodsky Chair and Associate Professor of Professor of Biological Sciences Biological Sciences University of Pittsburgh (Committee Member) (Committee Member) ________________________________ Dr. Philip Reeder Dean, Bayer School of Natural and Environmental Science iii ABSTRACT THE ROLE OF B-TYPE PHOSPHOLIPASES IN S. -

Metabolic Regulation by Lipid Activated Receptors by Maxwell A
Metabolic Regulation by Lipid Activated Receptors By Maxwell A Ruby A dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy In Molecular & Biochemical Nutrition In the Graduate Division Of the University of California, Berkeley Committee in charge: Professor Marc K. Hellerstein, Chair Professor Ronald M. Krauss Professor George A. Brooks Professor Andreas Stahl Fall 2010 Abstract Metabolic Regulation by Lipid Activated Receptors By Maxwell Alexander Ruby Doctor of Philosophy in Molecular & Biochemical Nutrition University of California, Berkeley Professor Marc K. Hellerstein, Chair Obesity and related metabolic disorders have reached epidemic levels with dire public health consequences. Efforts to stem the tide focus on behavioral and pharmacological interventions. Several hypolipidemic pharmaceutical agents target endogenous lipid receptors, including the peroxisomal proliferator activated receptor α (PPAR α) and cannabinoid receptor 1 (CB1). To further the understanding of these clinically relevant receptors, we elucidated the biochemical basis of PPAR α activation by lipoprotein lipolysis products and the metabolic and transcriptional responses to elevated endocannabinoid signaling. PPAR α is activated by fatty acids and their derivatives in vitro. While several specific pathways have been implicated in the generation of PPAR α ligands, we focused on lipoprotein lipase mediated lipolysis of triglyceride rich lipoproteins. Fatty acids activated PPAR α similarly to VLDL lipolytic products. Unbound fatty acid concentration determined the extent of PPAR α activation. Lipolysis of VLDL, but not physiological unbound fatty acid concentrations, created the fatty acid uptake necessary to stimulate PPAR α. Consistent with a role for vascular lipases in the activation of PPAR α, administration of a lipase inhibitor (p-407) prevented PPAR α dependent induction of target genes in fasted mice. -

Genetic and Molecular Analysis of Two New Loci Controlling Flowering in Garden Pea
Genetic and molecular analysis of two new loci controlling flowering in garden pea By A S M Mainul Hasan School of Natural Sciences Submitted in fulfilment of the requirements for the degree of Doctor of Philosophy University of Tasmania, July 2018 Declaration of originality This thesis contains no material which has been accepted for a degree or diploma by the University or any other institution, except by way of background information and duly acknowledge in the thesis, and to the best of my knowledge and belief no material previously published or written by another person except where due acknowledgement is made in the text of the thesis, nor does the thesis contain any material that infringes copyright. Authority of access This thesis may be made available for loan. Copying and communication of any part of this thesis is prohibited for two years from the date this statement was signed; after that time limited copying and communication is permitted in accordance with the Copyright Act 1968. Date: 6-07-2018 A S M Mainul Hasan i Abstract Flowering is one of the key developmental process associated with the life cycle of plant and it is regulated by different environmental factors and endogenous cues. In the model species Arabidopsis thaliana a mobile protein, FLOWERING LOCUS T (FT) plays central role to mediate flowering time and expression of FT is regulated by photoperiod. While flowering mechanisms are well-understood in A. thaliana, knowledge about this process is limited in legume (family Fabaceae) which are the second major group of crops after cereals in satisfying the global demand for food and fodder. -

Recombinant Human Carboxylesterase 1/CES1 Protein Catalog Number: ATGP3862
Recombinant human Carboxylesterase 1/CES1 protein Catalog Number: ATGP3862 PRODUCT INPORMATION Expression system Baculovirus Domain 19-568aa UniProt No. P23141 NCBI Accession No. NP_001020366 Alternative Names Liver carboxylesterase 1 isoform a, CES1, ACAT, CE-1, CEH, CES2, hCE-1, HMSE, HMSE1, PCE-1, REH, SES1, TGH PRODUCT SPECIFICATION Molecular Weight 61.7 kDa (559aa) Concentration 0.5mg/ml (determined by absorbance at 280nm) Formulation Liquid in. 25mM Sodium Acetate (pH 4.0) containing 10% glycerol, 0.1M NaCl, 0.1mM PMSF Purity > 90% by SDS-PAGE Endotoxin level < 1 EU per 1ug of protein (determined by LAL method) Biological Activity Specific activity is > 700pmol/min/ug and is defined as the amount of enzyme that hydrolyze 1pmole of p- nitrophenyl acetate to pnitrophenol per minute at pH 7.5 at 37C. Tag His-Tag Application Enzyme Activity,SDS-PAGE Storage Condition Can be stored at +2C to +8C for 1 week. For long term storage, aliquot and store at -20C to -80C. Avoid repeated freezing and thawing cycles. BACKGROUND 1 Recombinant human Carboxylesterase 1/CES1 protein Catalog Number: ATGP3862 Description Carboxylesterase 1, also known as liver carboxylesterase 1 isoform a, is a member of a large family of carboxylesterases. It is also part of the alpha/beta fold hydrolase family. Carboxylesterases hydrolyze long-chain fatty acid esters and thioesters. The resulting carboxylates are then often conjugated by other enzymes to increase solubility and eventually excreted. Also, this enzyme may play a role in detoxification in the lung and/or protection of the central nervous system from ester or amide compounds. -

A Middle-Aged Enzyme Still in Its Prime: Recent Advances in the Field of Cutinases
catalysts Review A Middle-Aged Enzyme Still in Its Prime: Recent Advances in the Field of Cutinases Efstratios Nikolaivits 1,† , Maria Kanelli 1,†, Maria Dimarogona 2 and Evangelos Topakas 1,* 1 IndBioCat Group, Biotechnology Laboratory, School of Chemical Engineering, National Technical University of Athens, 9 Heroon Polytechniou Str., Zographou Campus, 15780 Athens, Greece; [email protected] (E.N.); [email protected] (M.K.) 2 Department of Chemical Engineering, University of Patras, 26504 Patras, Greece; [email protected] * Correspondence: [email protected]; Tel.: +30-210-772-3264 † These authors have contributed equally in this work. Received: 7 November 2018; Accepted: 27 November 2018; Published: 3 December 2018 Abstract: Cutinases are α/β hydrolases, and their role in nature is the degradation of cutin. Such enzymes are usually produced by phytopathogenic microorganisms in order to penetrate their hosts. The first focused studies on cutinases started around 50 years ago. Since then, numerous cutinases have been isolated and characterized, aiming at the elucidation of their structure–function relations. Our deeper understanding of cutinases determines the applications by which they could be utilized; from food processing and detergents, to ester synthesis and polymerizations. However, cutinases are mainly efficient in the degradation of polyesters, a natural function. Therefore, these enzymes have been successfully applied for the biodegradation of plastics, as well as for the delicate superficial hydrolysis of polymeric materials prior to their functionalization. Even though research on this family of enzymes essentially began five decades ago, they are still involved in many reports; novel enzymes are being discovered, and new fields of applications arise, leading to numerous related publications per year.