Over-Expression and Characterization of a Glyoxalase 2 Like Enzyme
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Fruit Ripening and Storage
OPEN Citation: Horticulture Research (2014) 1, 6; doi:10.1038/hortres.2014.6 ß 2014 Nanjing Agricultural University All rights reserved 2052-7276/14 www.nature.com/hortres ARTICLE Dynamic changes in proteins during apple (Malus x domestica) fruit ripening and storage Yun Shi1, Li Jiang1, Li Zhang2, Ruoyi Kang1 and Zhifang Yu1 A proteomic study, using two-dimensional polyacrylamide gel electrophoresis and matrix-assisted laser desorption/ionization time-of-flight/time-of-flight, was conducted in apple fruit (cv. ‘Golden Delicious’) starting at 10 days prior to harvest through 50 days in storage. Total protein was extracted using a phenol/sodium dodecyl sulfate protocol. More than 400 protein spots were detected in each gel and 55 differentially expressed proteins (p,0.05) were subjected to matrix-assisted laser desorption/ionization time-of-flight/ time-of-flight analysis. Fifty-three of these proteins were finally identified using an apple expressed sequence tag database downloaded from Genome Database for Rosaceae and placed into six categories. The categories and the percentage of proteins placed in each category were stress response and defense (49.0%), energy and metabolism (34.0%), fruit ripening and senescence (5.6%), signal transduction (3.8%), cell structure (3.8%) and protein synthesis (3.8%). Proteins involved in several multiple metabolic pathways, including glycolysis, pentose–phosphate pathway, anti-oxidative systems, photosynthesis and cell wall synthesis, were downregulated, especially during the climacteric burst in respiration and during the senescent stages of fruit development. Proteins classified as allergens or involved in cell wall degradation were upregulated during the ripening process. Some protein spots exhibited a mixed pattern (increasing to maximal abundance followed by a decrease), such as 1-aminocyclopropane-1-carboxylate oxidase, L-ascorbate peroxidase and abscisic acid response proteins. -

Genome-Wide Analysis of Glyoxalase-Like Gene Families in Grape
Li et al. BMC Genomics (2019) 20:362 https://doi.org/10.1186/s12864-019-5733-y RESEARCHARTICLE Open Access Genome-wide analysis of glyoxalase-like gene families in grape (Vitis vinifera L.) and their expression profiling in response to downy mildew infection Tiemei Li1,2,3, Xin Cheng1,2,3, Yuting Wang1,2,3, Xiao Yin1,2,3, Zhiqian Li1,2,3, Ruiqi Liu1,2,3, Guotian Liu1,2,3, Yuejin Wang1,2,3 and Yan Xu1,2,3* Abstract Background: The glyoxalase system usually comprises two enzymes, glyoxalase I (GLYI) and glyoxalase II (GLYII). This system converts cytotoxic methylglyoxal (MG) into non-toxic D-lactate in the presence of reduced glutathione (GSH) in two enzymatic steps. Recently, a novel type of glyoxalase III (GLYIII) activity has observed in Escherichia coli that can detoxify MG into D-lactate directly, in one step, without a cofactor. Investigation of the glyoxalase enzymes of a number of plant species shows the importance of their roles in response both to abiotic and to biotic stresses. Until now, glyoxalase gene families have been identified in the genomes of four plants, Arabidopsis, Oryza sativa, Glycine max and Medicago truncatula but no similar study has been done with the grapevine Vitis vinifera L. Results: In this study, four GLYI-like,twoGLYII-like and three GLYIII-like genesareidentifiedfromthegenomedatabaseof grape. All these genes were analysed in detail, including their chromosomal locations, phylogenetic relationships, exon-intron distributions, protein domain organisations and the presence of conserved binding sites. Using quantitative real-time PCR analysis (qRT-PCR), the expression profiles of these geneswereanalysedindifferent tissues of grape, and also when under infection stress from downy mildew (Plasmopara viticola). -

Supplementary Materials
Supplementary Materials COMPARATIVE ANALYSIS OF THE TRANSCRIPTOME, PROTEOME AND miRNA PROFILE OF KUPFFER CELLS AND MONOCYTES Andrey Elchaninov1,3*, Anastasiya Lokhonina1,3, Maria Nikitina2, Polina Vishnyakova1,3, Andrey Makarov1, Irina Arutyunyan1, Anastasiya Poltavets1, Evgeniya Kananykhina2, Sergey Kovalchuk4, Evgeny Karpulevich5,6, Galina Bolshakova2, Gennady Sukhikh1, Timur Fatkhudinov2,3 1 Laboratory of Regenerative Medicine, National Medical Research Center for Obstetrics, Gynecology and Perinatology Named after Academician V.I. Kulakov of Ministry of Healthcare of Russian Federation, Moscow, Russia 2 Laboratory of Growth and Development, Scientific Research Institute of Human Morphology, Moscow, Russia 3 Histology Department, Medical Institute, Peoples' Friendship University of Russia, Moscow, Russia 4 Laboratory of Bioinformatic methods for Combinatorial Chemistry and Biology, Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow, Russia 5 Information Systems Department, Ivannikov Institute for System Programming of the Russian Academy of Sciences, Moscow, Russia 6 Genome Engineering Laboratory, Moscow Institute of Physics and Technology, Dolgoprudny, Moscow Region, Russia Figure S1. Flow cytometry analysis of unsorted blood sample. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S2. Flow cytometry analysis of unsorted liver stromal cells. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S3. MiRNAs expression analysis in monocytes and Kupffer cells. Full-length of heatmaps are presented. -
Supplementary File 1
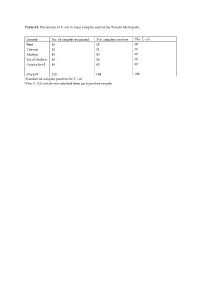
Table S1. Prevalence of E. coli in meat samples sold at the Tamale Metropolis. Sample No. of samples examined aNo. samples positive bNo. E. coli Beef 45 39 39 Chevon 45 34 34 Mutton 45 40 40 Local chicken 45 36 36 Guinea fowl 45 40 40 Overall 225 189 189 aNumber of samples positive for E. coli. bOne E. Coli isolate was selected from each positive sample. Table S2. A table showing the eBURST (Based Upon Related Sequence Types) analyses of the study sequence types with global curated STs in Escherichia PubMLST database. MLST (Isolate) Type of clone Closet global ancestry Source sequence type (ST) ST69 (SG6) Similar a ST69 Animal (Food), Human ST155 (SLC2, Similar ST155 Animal (Food), Human, TLC13, CM4) Environment ST297 (TLC1) Similar ST297 Human ST1727 (NC3) Similar ST1727 Human ST44 (AC1) Single-Locus Variant ST10, ST752 Animal (Food), (SLV) b Human ST469 (CC6) Single-Locus Variant ST162 Food (SLV) ST540 (AB1, Single-Locus Variant ST4093 Human TG1) (SLV) ST1141 (NM11) Single-Locus Variant ST10, ST744 Animal (Food), (SLV) Human ST7473 (NB12) Single-Locus Variant ST10 Animal (Food), (SLV) Human ST6646 (CB1) Satellite c None - ST7483 (NB12) Satellite None - a Similar: study isolate was similar to a global curated known sequence type. b Single-Locus Variant (SLV): study isolate only shared similarity with global curated known sequence types that differed in one allelic gene. c Satellite: study isolate as a distantly related and did not shared any similarity with global curated known sequence types. Table S3. In silico identification and characterization of conserved stress response mechanisms in the E. -

The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease Jonathan Z
REVIEW pubs.acs.org/CR The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease Jonathan Z. Long* and Benjamin F. Cravatt* The Skaggs Institute for Chemical Biology and Department of Chemical Physiology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, California 92037, United States CONTENTS 2.4. Other Phospholipases 6034 1. Introduction 6023 2.4.1. LIPG (Endothelial Lipase) 6034 2. Small-Molecule Hydrolases 6023 2.4.2. PLA1A (Phosphatidylserine-Specific 2.1. Intracellular Neutral Lipases 6023 PLA1) 6035 2.1.1. LIPE (Hormone-Sensitive Lipase) 6024 2.4.3. LIPH and LIPI (Phosphatidic Acid-Specific 2.1.2. PNPLA2 (Adipose Triglyceride Lipase) 6024 PLA1R and β) 6035 2.1.3. MGLL (Monoacylglycerol Lipase) 6025 2.4.4. PLB1 (Phospholipase B) 6035 2.1.4. DAGLA and DAGLB (Diacylglycerol Lipase 2.4.5. DDHD1 and DDHD2 (DDHD Domain R and β) 6026 Containing 1 and 2) 6035 2.1.5. CES3 (Carboxylesterase 3) 6026 2.4.6. ABHD4 (Alpha/Beta Hydrolase Domain 2.1.6. AADACL1 (Arylacetamide Deacetylase-like 1) 6026 Containing 4) 6036 2.1.7. ABHD6 (Alpha/Beta Hydrolase Domain 2.5. Small-Molecule Amidases 6036 Containing 6) 6027 2.5.1. FAAH and FAAH2 (Fatty Acid Amide 2.1.8. ABHD12 (Alpha/Beta Hydrolase Domain Hydrolase and FAAH2) 6036 Containing 12) 6027 2.5.2. AFMID (Arylformamidase) 6037 2.2. Extracellular Neutral Lipases 6027 2.6. Acyl-CoA Hydrolases 6037 2.2.1. PNLIP (Pancreatic Lipase) 6028 2.6.1. FASN (Fatty Acid Synthase) 6037 2.2.2. PNLIPRP1 and PNLIPR2 (Pancreatic 2.6.2. -

Isolation and Characterization of the Prolyl Aminopeptidase Gene (Pap) from Aeromonas Sobria: Comparison with the Bacillus Coagulans Enzyme1
J. Biochem. 116, 818-825 (1994) Isolation and Characterization of the Prolyl Aminopeptidase Gene (pap) from Aeromonas sobria: Comparison with the Bacillus coagulans Enzyme1 Ana Kitazono,* Atsuko Kitano,* Daisuke Tsuru,•õ and Tadashi Yoshimoto*,2 *School of Pharmaceutical Sciences , Nagasaki University, 1-14 Bunkyo-machi, Nagasaki, Nagasaki 852; and •õ Department of Applied Microbiology, Kumamoto Institute of Technology, 4-22-1 Ikeda, Kumamoto, Kumamoto 860 Received for publication, May 16, 1994 The Aeromonas sobria pap gene encoding prolyl aminopeptidase (PAP) was cloned. It consists of 425 codons and encodes a homotetrameric enzyme of 205kDa. The purified enzyme showed an almost absolute specificity for amino-terminal proline. Proline and hydroxyproline residues from many peptide and amide substrates could be easily removed, while no activity was detected for substrates having other amino terminals. The enzyme was very similar to that from Bacillus coagulans in many aspects, such as the strong inhibition caused by PCMB and the weak or no inhibition caused by DFP and chelators, respectively. However, these enzymes show only 15% identity in their amino acid sequences. Differences were also observed in their molecular weight, stability and activity toward some peptide substrates. When aligning the deduced amino acid sequence with known sequences from other microorganisms, conserved sequences were found at the amino-terminal region; the significance of these conserved regions is discussed. Based on the results of this work, and on the studies available to date, the occurrence of at least two types of PAPs is postulated. One group would be formed by the Bacillus, Neisseria, and Lactobacillus enzymes, and the other by enzymes such as the Aeromonas PAP. -

Supplementary Information 2 to Accompany
1 Supplementary Information 2 to accompany 3 Sulfur-oxidizing symbionts without canonical genes for autotrophic CO2 fixation 4 Brandon K. B. Seah*1,7, Chakkiath Paul Antony1,8, Bruno Huettel2, Jan Zarzycki3, Lennart 5 Schada von Borzyskowski3, Tobias J. Erb3, Angela Kouris4, Manuel Kleiner5, Manuel 6 Liebeke1, Nicole Dubilier1,6, Harald R. Gruber-Vodicka1 7 1 Max Planck Institute for Marine Microbiology, Celsiusstraße 1, 28359 Bremen, Germany 8 2 Max Planck Genome Centre Cologne, Max Planck Institute for Plant Breeding Research, 9 Carl-von-Linné-Weg 10, 50829 Cologne, Germany 10 3 Max Planck Institute for Terrestrial Microbiology, Karl-von-Frisch-Str. 10, 35043 Marburg, 11 Germany 12 4 Energy Bioengineering and Geomicrobiology Group, University of Calgary, 2500 13 University Drive Northwest, Calgary, Alberta T2N 1N4, Canada 14 5 Department of Plant and Microbial Biology, North Carolina State University, Raleigh 15 27695, North Carolina, United States of America 16 6 MARUM, Center for Marine Environmental Sciences, University of Bremen, 28359 17 Bremen, Germany 18 7 Current address: Max Planck Institute for Developmental Biology, Max-Planck-Ring 5, 19 72076 Tübingen, Germany 20 8 Current address: Red Sea Research Center, Biological and Environmental Sciences and 21 Engineering (BESE) Division, King Abdullah University of Science and Technology 22 (KAUST), Thuwal 23955, Kingdom of Saudi Arabia 23 * Corresponding author 1 24 Supplementary Materials and Methods 25 Metabolite extraction and identification 26 Kentrophoros sp. H was collected on Elba in 2014 for metabolomics (Supplementary Table 27 7). Samples were fixed in 1 mL cold methanol (HPLC-grade, Sigma-Aldrich) and stored at - 28 20°C until use. -

P-Glycoprotein, CYP3A, and Plasma Carboxylesterase Determine Brain and Blood Disposition of the Mtor Inhibitor Everolimus (Afinitor) in Mice
Published OnlineFirst April 11, 2014; DOI: 10.1158/1078-0432.CCR-13-1759 Clinical Cancer Cancer Therapy: Preclinical Research P-Glycoprotein, CYP3A, and Plasma Carboxylesterase Determine Brain and Blood Disposition of the mTOR Inhibitor Everolimus (Afinitor) in Mice Seng Chuan Tang1, Rolf W. Sparidans3, Ka Lei Cheung4, Tatsuki Fukami5, Selvi Durmus1, Els Wagenaar1, Tsuyoshi Yokoi5, Bart J.M. van Vlijmen4, Jos H. Beijnen2,3, and Alfred H. Schinkel1 Abstract Purpose: To clarify the role of ABCB1, ABCG2, and CYP3A in blood and brain exposure of everolimus using knockout mouse models. À À À À À À À À Experimental Design: We used wild-type, Abcb1a/1b / , Abcg2 / , Abcb1a/1b;Abcg2 / , and Cyp3a / mice to study everolimus oral bioavailability and brain accumulation. Results: Following everolimus administration, brain concentrations and brain-to-liver ratios were À À À À À À substantially increased in Abcb1a/1b / and Abcb1a/1b;Abcg2 / , but not Abcg2 / mice. The fraction of everolimus located in the plasma compartment was highly increased in all knockout strains. In vitro, everolimus was rapidly degraded in wild-type but not knockout plasma. Carboxylesterase 1c (Ces1c), a plasma carboxylesterase gene, was highly upregulated (80-fold) in the liver of knockout mice relative to wild-type mice, and plasma Ces1c likely protected everolimus from degradation by binding and stabilizing it. This binding was prevented by preincubation with the carboxylesterase inhibitor BNPP. In vivo knockdown experiments confirmed the involvement of Ces1c in everolimus stabilization. Everolimus also markedly inhibited the hydrolysis of irinotecan and p-nitrophenyl acetate by mouse plasma carboxylesterase À À and recombinant human CES2, respectively. -

Electronic Supplementary Material (ESI) for Metallomics
Electronic Supplementary Material (ESI) for Metallomics. This journal is © The Royal Society of Chemistry 2018 Uniprot Entry name Gene names Protein names Predicted Pattern Number of Iron role EC number Subcellular Membrane Involvement in disease Gene ontology (biological process) Id iron ions location associated 1 P46952 3HAO_HUMAN HAAO 3-hydroxyanthranilate 3,4- H47-E53-H91 1 Fe cation Catalytic 1.13.11.6 Cytoplasm No NAD biosynthetic process [GO:0009435]; neuron cellular homeostasis dioxygenase (EC 1.13.11.6) (3- [GO:0070050]; quinolinate biosynthetic process [GO:0019805]; response to hydroxyanthranilate oxygenase) cadmium ion [GO:0046686]; response to zinc ion [GO:0010043]; tryptophan (3-HAO) (3-hydroxyanthranilic catabolic process [GO:0006569] acid dioxygenase) (HAD) 2 O00767 ACOD_HUMAN SCD Acyl-CoA desaturase (EC H120-H125-H157-H161; 2 Fe cations Catalytic 1.14.19.1 Endoplasmic Yes long-chain fatty-acyl-CoA biosynthetic process [GO:0035338]; unsaturated fatty 1.14.19.1) (Delta(9)-desaturase) H160-H269-H298-H302 reticulum acid biosynthetic process [GO:0006636] (Delta-9 desaturase) (Fatty acid desaturase) (Stearoyl-CoA desaturase) (hSCD1) 3 Q6ZNF0 ACP7_HUMAN ACP7 PAPL PAPL1 Acid phosphatase type 7 (EC D141-D170-Y173-H335 1 Fe cation Catalytic 3.1.3.2 Extracellular No 3.1.3.2) (Purple acid space phosphatase long form) 4 Q96SZ5 AEDO_HUMAN ADO C10orf22 2-aminoethanethiol dioxygenase H112-H114-H193 1 Fe cation Catalytic 1.13.11.19 Unknown No oxidation-reduction process [GO:0055114]; sulfur amino acid catabolic process (EC 1.13.11.19) (Cysteamine -

Hydrogen Sulfide Metabolite, Sodium Thiosulfate
International Journal of Molecular Sciences Review Hydrogen Sulfide Metabolite, Sodium Thiosulfate: Clinical Applications and Underlying Molecular Mechanisms Max Y. Zhang 1,2, George J. Dugbartey 1,2,3, Smriti Juriasingani 1,3 and Alp Sener 1,2,3,4,* 1 Matthew Mailing Center for Translational Transplant Studies, London Health Sciences Center, Western University, London, ON N6A 5A5, Canada; [email protected] (M.Y.Z.); [email protected] (G.J.D.); [email protected] (S.J.) 2 London Health Sciences Center, Multi-Organ Transplant Program, Western University, London, ON N6A 5A5, Canada 3 London Health Sciences Center, Department of Surgery, Division of Urology, Western University, London, ON N6A 5A5, Canada 4 Department of Microbiology & Immunology, Schulich School of Medicine & Dentistry, University of Western Ontario, London, ON N6A 3K7, Canada * Correspondence: [email protected]; Tel.: +1(519) 6633352 Abstract: Thiosulfate in the form of sodium thiosulfate (STS) is a major oxidation product of hydrogen sulfide (H2S), an endogenous signaling molecule and the third member of the gasotransmitter family. STS is currently used in the clinical treatment of acute cyanide poisoning, cisplatin toxicities in cancer therapy, and calciphylaxis in dialysis patients. Burgeoning evidence show that STS has antioxidant and anti-inflammatory properties, making it a potential therapeutic candidate molecule that can target multiple molecular pathways in various diseases and drug-induced toxicities. This review Citation: Zhang, M.Y.; Dugbartey, discusses the biochemical and molecular pathways in the generation of STS from H2S, its clinical G.J.; Juriasingani, S.; Sener, A. usefulness, and potential clinical applications, as well as the molecular mechanisms underlying these Hydrogen Sulfide Metabolite, clinical applications and a future perspective in kidney transplantation. -

Recombinant Human Glyoxalase I Catalog Number: 4959-GL
Recombinant Human Glyoxalase I Catalog Number: 4959-GL DESCRIPTION Source E. coliderived Ala2Met184, with an Nterminal Met and 6His tag Accession # NP_006699 Nterminal Sequence Met Analysis Predicted Molecular 22 kDa Mass SPECIFICATIONS SDSPAGE 25 kDa, reducing conditions Activity Measured by its ability to catalyze the formation of SDlactoylglutathione from the hemimercaptal adduct that forms spontaneously between methylglyoxal and reduced glutathione. The specific activity is >100 nmol/min/µg, as measured under the described conditions. Endotoxin Level <1.0 EU per 1 μg of the protein by the LAL method. Purity >85%, by SDSPAGE under reducing conditions and visualized by silver stain. Formulation Lyophilized from a 0.2 μm filtered solution in TrisHCl and DTT. See Certificate of Analysis for details. Activity Assay Protocol Materials l Assay Buffer: 0.1 M Sodium Phosphate, pH 7.0 l Recombinant Human Glyoxalase I (rhGlyoxalase I) (Catalog # 4959GL) l Glutathione, Reduced (GSH) (Amresco, Catalog # 0399) l Methylglyoxal solution, 40% (Sigma, Catalog # M0252) l 96well Clear UV Plate (Costar, Catalog # 3635) l Plate Reader (Model: SpectraMax Plus by Molecular Devices) or equivalent Assay 1. Prepare 100 mM GSH in deionized water. Note: Prepare fresh. 2. Dilute 40% (6.48 M) Methylglyoxal solution to 100 mM in Assay Buffer. Note: Prepare fresh. 3. Combine 1420 µL Assay Buffer, 40 µL 100 mM GSH, and 40 µL 100 mM Methylglyoxal to make the Substrate Mixture. 4. Incubate at room temperature for 15 minutes. 5. Dilute rhGlyoxalase I to 0.4 ng/µL in Assay Buffer. -

Springer Handbook of Enzymes
Dietmar Schomburg and Ida Schomburg (Eds.) Springer Handbook of Enzymes Volume 25 Class 1 • Oxidoreductases X EC 1.9-1.13 co edited by Antje Chang Second Edition 4y Springer Index of Recommended Enzyme Names EC-No. Recommended Name Page 1.13.11.50 acetylacetone-cleaving enzyme 673 1.10.3.4 o-aminophenol oxidase 149 1.13.12.12 apo-/?-carotenoid-14',13'-dioxygenase 732 1.13.11.34 arachidonate 5-lipoxygenase 591 1.13.11.40 arachidonate 8-lipoxygenase 627 1.13.11.31 arachidonate 12-lipoxygenase 568 1.13.11.33 arachidonate 15-lipoxygenase 585 1.13.12.1 arginine 2-monooxygenase 675 1.13.11.13 ascorbate 2,3-dioxygenase 491 1.10.2.1 L-ascorbate-cytochrome-b5 reductase 79 1.10.3.3 L-ascorbate oxidase 134 1.11.1.11 L-ascorbate peroxidase 257 1.13.99.2 benzoate 1,2-dioxygenase (transferred to EC 1.14.12.10) 740 1.13.11.39 biphenyl-2,3-diol 1,2-dioxygenase 618 1.13.11.22 caffeate 3,4-dioxygenase 531 1.13.11.16 3-carboxyethylcatechol 2,3-dioxygenase 505 1.13.11.21 p-carotene 15,15'-dioxygenase (transferred to EC 1.14.99.36) 530 1.11.1.6 catalase 194 1.13.11.1 catechol 1,2-dioxygenase 382 1.13.11.2 catechol 2,3-dioxygenase 395 1.10.3.1 catechol oxidase 105 1.13.11.36 chloridazon-catechol dioxygenase 607 1.11.1.10 chloride peroxidase 245 1.13.11.49 chlorite O2-lyase 670 1.13.99.4 4-chlorophenylacetate 3,4-dioxygenase (transferred to EC 1.14.12.9) .