Isolation and Characterization of the Prolyl Aminopeptidase Gene (Pap) from Aeromonas Sobria: Comparison with the Bacillus Coagulans Enzyme1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Characterisation of Aspergillus Niger Prolyl Aminopeptidase
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Wageningen University & Research Publications Mol Gen Genomics (2005) 272: 673–679 DOI 10.1007/s00438-004-1094-5 ORIGINAL PAPER Danie¨lle E. J. W. Basten Æ Antoine P. H. A. Moers Albert J. J. van. Ooyen Æ Peter J. Schaap Characterisation of Aspergillus niger prolyl aminopeptidase Received: 29 April 2004 / Accepted: 16 November 2004 / Published online: 15 January 2005 Ó Springer-Verlag 2005 Abstract We have cloned a gene (papA) that encodes a ases and tripeptidases and finally by carboxypeptidases prolyl aminopeptidase from Aspergillus niger. Homolo- and aminopeptidases. The turnover of proteins by pro- gous genes are present in the genomes of the Eurotiales teases provides a ready pool of amino acids as precur- A. nidulans, A. fumigatus and Talaromyces emersonii, sors for the synthesis of new proteins (Bennet and Klich but the gene is not present in the genome of the yeast 1992). Saccharomyces cerevisiae. Cell extracts of strains over- Proteases normally do not hydrolyse bonds adjacent expressing the gene under the control of its own pro- to proline residues. Instead a specialised group of en- moter showed a fourfold to sixfold increase in prolyl zymes has evolved that hydrolyses these bonds. Their aminopeptidase activity, but no change in phenylalanine activity depends on both the isomeric state of the proline or leucine aminopeptidase activity. The overexpressed residue and its position in the peptide chain (Vanhoof enzyme was subsequently purified and characterised. et al. 1995; Cunningham and O’Connor 1997). Proline The enzyme specifically removes N-terminal proline and aminopeptidases (Pap, prolyl iminopeptidase, EC hydroxyproline residues from peptides. -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of -

Serine Proteases with Altered Sensitivity to Activity-Modulating
(19) & (11) EP 2 045 321 A2 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 08.04.2009 Bulletin 2009/15 C12N 9/00 (2006.01) C12N 15/00 (2006.01) C12Q 1/37 (2006.01) (21) Application number: 09150549.5 (22) Date of filing: 26.05.2006 (84) Designated Contracting States: • Haupts, Ulrich AT BE BG CH CY CZ DE DK EE ES FI FR GB GR 51519 Odenthal (DE) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • Coco, Wayne SK TR 50737 Köln (DE) •Tebbe, Jan (30) Priority: 27.05.2005 EP 05104543 50733 Köln (DE) • Votsmeier, Christian (62) Document number(s) of the earlier application(s) in 50259 Pulheim (DE) accordance with Art. 76 EPC: • Scheidig, Andreas 06763303.2 / 1 883 696 50823 Köln (DE) (71) Applicant: Direvo Biotech AG (74) Representative: von Kreisler Selting Werner 50829 Köln (DE) Patentanwälte P.O. Box 10 22 41 (72) Inventors: 50462 Köln (DE) • Koltermann, André 82057 Icking (DE) Remarks: • Kettling, Ulrich This application was filed on 14-01-2009 as a 81477 München (DE) divisional application to the application mentioned under INID code 62. (54) Serine proteases with altered sensitivity to activity-modulating substances (57) The present invention provides variants of ser- screening of the library in the presence of one or several ine proteases of the S1 class with altered sensitivity to activity-modulating substances, selection of variants with one or more activity-modulating substances. A method altered sensitivity to one or several activity-modulating for the generation of such proteases is disclosed, com- substances and isolation of those polynucleotide se- prising the provision of a protease library encoding poly- quences that encode for the selected variants. -

Supplemental Methods
Supplemental Methods: Sample Collection Duplicate surface samples were collected from the Amazon River plume aboard the R/V Knorr in June 2010 (4 52.71’N, 51 21.59’W) during a period of high river discharge. The collection site (Station 10, 4° 52.71’N, 51° 21.59’W; S = 21.0; T = 29.6°C), located ~ 500 Km to the north of the Amazon River mouth, was characterized by the presence of coastal diatoms in the top 8 m of the water column. Sampling was conducted between 0700 and 0900 local time by gently impeller pumping (modified Rule 1800 submersible sump pump) surface water through 10 m of tygon tubing (3 cm) to the ship's deck where it then flowed through a 156 µm mesh into 20 L carboys. In the lab, cells were partitioned into two size fractions by sequential filtration (using a Masterflex peristaltic pump) of the pre-filtered seawater through a 2.0 µm pore-size, 142 mm diameter polycarbonate (PCTE) membrane filter (Sterlitech Corporation, Kent, CWA) and a 0.22 µm pore-size, 142 mm diameter Supor membrane filter (Pall, Port Washington, NY). Metagenomic and non-selective metatranscriptomic analyses were conducted on both pore-size filters; poly(A)-selected (eukaryote-dominated) metatranscriptomic analyses were conducted only on the larger pore-size filter (2.0 µm pore-size). All filters were immediately submerged in RNAlater (Applied Biosystems, Austin, TX) in sterile 50 mL conical tubes, incubated at room temperature overnight and then stored at -80oC until extraction. Filtration and stabilization of each sample was completed within 30 min of water collection. -

Changes in the Sclerotinia Sclerotiorum Transcriptome During Infection of Brassica Napus
Seifbarghi et al. BMC Genomics (2017) 18:266 DOI 10.1186/s12864-017-3642-5 RESEARCHARTICLE Open Access Changes in the Sclerotinia sclerotiorum transcriptome during infection of Brassica napus Shirin Seifbarghi1,2, M. Hossein Borhan1, Yangdou Wei2, Cathy Coutu1, Stephen J. Robinson1 and Dwayne D. Hegedus1,3* Abstract Background: Sclerotinia sclerotiorum causes stem rot in Brassica napus, which leads to lodging and severe yield losses. Although recent studies have explored significant progress in the characterization of individual S. sclerotiorum pathogenicity factors, a gap exists in profiling gene expression throughout the course of S. sclerotiorum infection on a host plant. In this study, RNA-Seq analysis was performed with focus on the events occurring through the early (1 h) to the middle (48 h) stages of infection. Results: Transcript analysis revealed the temporal pattern and amplitude of the deployment of genes associated with aspects of pathogenicity or virulence during the course of S. sclerotiorum infection on Brassica napus. These genes were categorized into eight functional groups: hydrolytic enzymes, secondary metabolites, detoxification, signaling, development, secreted effectors, oxalic acid and reactive oxygen species production. The induction patterns of nearly all of these genes agreed with their predicted functions. Principal component analysis delineated gene expression patterns that signified transitions between pathogenic phases, namely host penetration, ramification and necrotic stages, and provided evidence for the occurrence of a brief biotrophic phase soon after host penetration. Conclusions: The current observations support the notion that S. sclerotiorum deploys an array of factors and complex strategies to facilitate host colonization and mitigate host defenses. This investigation provides a broad overview of the sequential expression of virulence/pathogenicity-associated genes during infection of B. -

The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease Jonathan Z
REVIEW pubs.acs.org/CR The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease Jonathan Z. Long* and Benjamin F. Cravatt* The Skaggs Institute for Chemical Biology and Department of Chemical Physiology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, California 92037, United States CONTENTS 2.4. Other Phospholipases 6034 1. Introduction 6023 2.4.1. LIPG (Endothelial Lipase) 6034 2. Small-Molecule Hydrolases 6023 2.4.2. PLA1A (Phosphatidylserine-Specific 2.1. Intracellular Neutral Lipases 6023 PLA1) 6035 2.1.1. LIPE (Hormone-Sensitive Lipase) 6024 2.4.3. LIPH and LIPI (Phosphatidic Acid-Specific 2.1.2. PNPLA2 (Adipose Triglyceride Lipase) 6024 PLA1R and β) 6035 2.1.3. MGLL (Monoacylglycerol Lipase) 6025 2.4.4. PLB1 (Phospholipase B) 6035 2.1.4. DAGLA and DAGLB (Diacylglycerol Lipase 2.4.5. DDHD1 and DDHD2 (DDHD Domain R and β) 6026 Containing 1 and 2) 6035 2.1.5. CES3 (Carboxylesterase 3) 6026 2.4.6. ABHD4 (Alpha/Beta Hydrolase Domain 2.1.6. AADACL1 (Arylacetamide Deacetylase-like 1) 6026 Containing 4) 6036 2.1.7. ABHD6 (Alpha/Beta Hydrolase Domain 2.5. Small-Molecule Amidases 6036 Containing 6) 6027 2.5.1. FAAH and FAAH2 (Fatty Acid Amide 2.1.8. ABHD12 (Alpha/Beta Hydrolase Domain Hydrolase and FAAH2) 6036 Containing 12) 6027 2.5.2. AFMID (Arylformamidase) 6037 2.2. Extracellular Neutral Lipases 6027 2.6. Acyl-CoA Hydrolases 6037 2.2.1. PNLIP (Pancreatic Lipase) 6028 2.6.1. FASN (Fatty Acid Synthase) 6037 2.2.2. PNLIPRP1 and PNLIPR2 (Pancreatic 2.6.2. -

P-Glycoprotein, CYP3A, and Plasma Carboxylesterase Determine Brain and Blood Disposition of the Mtor Inhibitor Everolimus (Afinitor) in Mice
Published OnlineFirst April 11, 2014; DOI: 10.1158/1078-0432.CCR-13-1759 Clinical Cancer Cancer Therapy: Preclinical Research P-Glycoprotein, CYP3A, and Plasma Carboxylesterase Determine Brain and Blood Disposition of the mTOR Inhibitor Everolimus (Afinitor) in Mice Seng Chuan Tang1, Rolf W. Sparidans3, Ka Lei Cheung4, Tatsuki Fukami5, Selvi Durmus1, Els Wagenaar1, Tsuyoshi Yokoi5, Bart J.M. van Vlijmen4, Jos H. Beijnen2,3, and Alfred H. Schinkel1 Abstract Purpose: To clarify the role of ABCB1, ABCG2, and CYP3A in blood and brain exposure of everolimus using knockout mouse models. À À À À À À À À Experimental Design: We used wild-type, Abcb1a/1b / , Abcg2 / , Abcb1a/1b;Abcg2 / , and Cyp3a / mice to study everolimus oral bioavailability and brain accumulation. Results: Following everolimus administration, brain concentrations and brain-to-liver ratios were À À À À À À substantially increased in Abcb1a/1b / and Abcb1a/1b;Abcg2 / , but not Abcg2 / mice. The fraction of everolimus located in the plasma compartment was highly increased in all knockout strains. In vitro, everolimus was rapidly degraded in wild-type but not knockout plasma. Carboxylesterase 1c (Ces1c), a plasma carboxylesterase gene, was highly upregulated (80-fold) in the liver of knockout mice relative to wild-type mice, and plasma Ces1c likely protected everolimus from degradation by binding and stabilizing it. This binding was prevented by preincubation with the carboxylesterase inhibitor BNPP. In vivo knockdown experiments confirmed the involvement of Ces1c in everolimus stabilization. Everolimus also markedly inhibited the hydrolysis of irinotecan and p-nitrophenyl acetate by mouse plasma carboxylesterase À À and recombinant human CES2, respectively. -

Supplementary Table 3 Gene Microarray Analysis: PRL+E2 Vs
Supplementary Table 3 Gene microarray analysis: PRL+E2 vs. control ID1 Field1 ID Symbol Name M Fold P Value 69 15562 206115_at EGR3 early growth response 3 2,36 5,13 4,51E-06 56 41486 232231_at RUNX2 runt-related transcription factor 2 2,01 4,02 6,78E-07 41 36660 227404_s_at EGR1 early growth response 1 1,99 3,97 2,20E-04 396 54249 36711_at MAFF v-maf musculoaponeurotic fibrosarcoma oncogene homolog F 1,92 3,79 7,54E-04 (avian) 42 13670 204222_s_at GLIPR1 GLI pathogenesis-related 1 (glioma) 1,91 3,76 2,20E-04 65 11080 201631_s_at IER3 immediate early response 3 1,81 3,50 3,50E-06 101 36952 227697_at SOCS3 suppressor of cytokine signaling 3 1,76 3,38 4,71E-05 16 15514 206067_s_at WT1 Wilms tumor 1 1,74 3,34 1,87E-04 171 47873 238623_at NA NA 1,72 3,30 1,10E-04 600 14687 205239_at AREG amphiregulin (schwannoma-derived growth factor) 1,71 3,26 1,51E-03 256 36997 227742_at CLIC6 chloride intracellular channel 6 1,69 3,23 3,52E-04 14 15038 205590_at RASGRP1 RAS guanyl releasing protein 1 (calcium and DAG-regulated) 1,68 3,20 1,87E-04 55 33237 223961_s_at CISH cytokine inducible SH2-containing protein 1,67 3,19 6,49E-07 78 32152 222872_x_at OBFC2A oligonucleotide/oligosaccharide-binding fold containing 2A 1,66 3,15 1,23E-05 1969 32201 222921_s_at HEY2 hairy/enhancer-of-split related with YRPW motif 2 1,64 3,12 1,78E-02 122 13463 204015_s_at DUSP4 dual specificity phosphatase 4 1,61 3,06 5,97E-05 173 36466 227210_at NA NA 1,60 3,04 1,10E-04 117 40525 231270_at CA13 carbonic anhydrase XIII 1,59 3,02 5,62E-05 81 42339 233085_s_at OBFC2A oligonucleotide/oligosaccharide-binding -
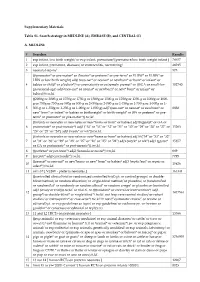
EMBASE (B), and CENTRAL (C) A. MEDLINE # Searches
Supplementary Materials Table S1. Search strategy in MEDLINE (A), EMBASE (B), and CENTRAL (C) A. MEDLINE # Searches Results 1 exp infant, low birth weight/ or exp infant, premature/ [premature/low birth weight infant ] 78657 2 exp infant, premature, diseases/ or enterocolitis, necrotizing/ 46015 3 neonatal sepsis/ 575 (((prematur* or pre-matur* or i?matur* or preterm* or pre-term* or VLBW* or ELBW* or LBW or low birth weight) adj6 (neo-nat* or neonat* or newborn* or born* or infant* or 4 babies or child* or p?ediatr*)) or prematurity or extremely premat* or ((SGA or small-for- 102740 gestational-age) adj6 (neo-nat* or neonat* or newborn* or new* born* or infant* or babies))).tw,kf. ((2000g or 2000-g or 1750g or 1750-g or 1500g or 1500-g or 1250g or 1250-g or 1000g or 1000- g or 750g or 750-g or 500g or 500-g or 2-000g or 2-000-g or 1-750g or 1-750-g or 1-500g or 1- 5 500-g or 1-250g or 1-250-g or 1-000g or 1-000-g) adj7 (neo-nat* or neonat* or newborn* or 8838 new* born* or infant* or babies or birthweight* or birth weight* or BW or preterm* or pre- term* or prematur* or pre-matur*)).tw,kf. ((infants or neonates or neo-nates or new*borns or born* or babies) adj18 (gestat* or GA or 6 postmenstr* or post-menstr*) adj3 ("34" or "33" or "32" or "31" or "30" or "29" or "28" or "27" or 15263 "26" or "25" or "24") adj3 (week* or wk*)).tw,kf. -

The Role of the B-Type Phospholipases in S. Cerevisiae: Function, Regulation, and Physiological Relevance in Lipid Homeostasis Beth a Surlow
Duquesne University Duquesne Scholarship Collection Electronic Theses and Dissertations Summer 2014 The Role of the B-Type Phospholipases in S. cerevisiae: Function, Regulation, and Physiological Relevance in Lipid Homeostasis Beth A Surlow Follow this and additional works at: https://dsc.duq.edu/etd Recommended Citation Surlow, B. (2014). The Role of the B-Type Phospholipases in S. cerevisiae: Function, Regulation, and Physiological Relevance in Lipid Homeostasis (Doctoral dissertation, Duquesne University). Retrieved from https://dsc.duq.edu/etd/1255 This Immediate Access is brought to you for free and open access by Duquesne Scholarship Collection. It has been accepted for inclusion in Electronic Theses and Dissertations by an authorized administrator of Duquesne Scholarship Collection. For more information, please contact [email protected]. THE ROLE OF B-TYPE PHOSPHOLIPASES IN S. CEREVISIAE: FUNCTION, REGULATION, AND PHYSIOLOGICAL RELEVANCE IN LIPID HOMEOSTASIS A Dissertation Submitted to the Bayer School of Natural and Environmental Sciences Duquesne University In partial fulfillment of the requirements for the degree of Doctor of Philosophy By Beth A. Surlow August 2014 Copyright by Beth A. Surlow 2014 THE ROLE OF B-TYPE PHOSPHOLIPASES IN S. CEREVISIAE: FUNCTION, REGULATION, AND PHYSIOLOGICAL RELEVANCE IN LIPID HOMEOSTASIS By Beth A. Surlow Approved June 5, 2014 ________________________________ ________________________________ Dr. Jana Patton-Vogt Dr. Philip Auron Associate Professor of Biological Professor of Biological Sciences Sciences (Committee Member) (Committee Chair) ________________________________ ________________________________ Dr. Joseph McCormick Dr. Jeffrey Brodsky Chair and Associate Professor of Professor of Biological Sciences Biological Sciences University of Pittsburgh (Committee Member) (Committee Member) ________________________________ Dr. Philip Reeder Dean, Bayer School of Natural and Environmental Science iii ABSTRACT THE ROLE OF B-TYPE PHOSPHOLIPASES IN S. -

Metabolic Regulation by Lipid Activated Receptors by Maxwell A
Metabolic Regulation by Lipid Activated Receptors By Maxwell A Ruby A dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy In Molecular & Biochemical Nutrition In the Graduate Division Of the University of California, Berkeley Committee in charge: Professor Marc K. Hellerstein, Chair Professor Ronald M. Krauss Professor George A. Brooks Professor Andreas Stahl Fall 2010 Abstract Metabolic Regulation by Lipid Activated Receptors By Maxwell Alexander Ruby Doctor of Philosophy in Molecular & Biochemical Nutrition University of California, Berkeley Professor Marc K. Hellerstein, Chair Obesity and related metabolic disorders have reached epidemic levels with dire public health consequences. Efforts to stem the tide focus on behavioral and pharmacological interventions. Several hypolipidemic pharmaceutical agents target endogenous lipid receptors, including the peroxisomal proliferator activated receptor α (PPAR α) and cannabinoid receptor 1 (CB1). To further the understanding of these clinically relevant receptors, we elucidated the biochemical basis of PPAR α activation by lipoprotein lipolysis products and the metabolic and transcriptional responses to elevated endocannabinoid signaling. PPAR α is activated by fatty acids and their derivatives in vitro. While several specific pathways have been implicated in the generation of PPAR α ligands, we focused on lipoprotein lipase mediated lipolysis of triglyceride rich lipoproteins. Fatty acids activated PPAR α similarly to VLDL lipolytic products. Unbound fatty acid concentration determined the extent of PPAR α activation. Lipolysis of VLDL, but not physiological unbound fatty acid concentrations, created the fatty acid uptake necessary to stimulate PPAR α. Consistent with a role for vascular lipases in the activation of PPAR α, administration of a lipase inhibitor (p-407) prevented PPAR α dependent induction of target genes in fasted mice. -

Genetic and Molecular Analysis of Two New Loci Controlling Flowering in Garden Pea
Genetic and molecular analysis of two new loci controlling flowering in garden pea By A S M Mainul Hasan School of Natural Sciences Submitted in fulfilment of the requirements for the degree of Doctor of Philosophy University of Tasmania, July 2018 Declaration of originality This thesis contains no material which has been accepted for a degree or diploma by the University or any other institution, except by way of background information and duly acknowledge in the thesis, and to the best of my knowledge and belief no material previously published or written by another person except where due acknowledgement is made in the text of the thesis, nor does the thesis contain any material that infringes copyright. Authority of access This thesis may be made available for loan. Copying and communication of any part of this thesis is prohibited for two years from the date this statement was signed; after that time limited copying and communication is permitted in accordance with the Copyright Act 1968. Date: 6-07-2018 A S M Mainul Hasan i Abstract Flowering is one of the key developmental process associated with the life cycle of plant and it is regulated by different environmental factors and endogenous cues. In the model species Arabidopsis thaliana a mobile protein, FLOWERING LOCUS T (FT) plays central role to mediate flowering time and expression of FT is regulated by photoperiod. While flowering mechanisms are well-understood in A. thaliana, knowledge about this process is limited in legume (family Fabaceae) which are the second major group of crops after cereals in satisfying the global demand for food and fodder.