Table SI. Downregulated Differentially Expressed Genes Identified in Lung Adenocarcinomas
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Single-Cell RNA Sequencing Demonstrates the Molecular and Cellular Reprogramming of Metastatic Lung Adenocarcinoma
ARTICLE https://doi.org/10.1038/s41467-020-16164-1 OPEN Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma Nayoung Kim 1,2,3,13, Hong Kwan Kim4,13, Kyungjong Lee 5,13, Yourae Hong 1,6, Jong Ho Cho4, Jung Won Choi7, Jung-Il Lee7, Yeon-Lim Suh8,BoMiKu9, Hye Hyeon Eum 1,2,3, Soyean Choi 1, Yoon-La Choi6,10,11, Je-Gun Joung1, Woong-Yang Park 1,2,6, Hyun Ae Jung12, Jong-Mu Sun12, Se-Hoon Lee12, ✉ ✉ Jin Seok Ahn12, Keunchil Park12, Myung-Ju Ahn 12 & Hae-Ock Lee 1,2,3,6 1234567890():,; Advanced metastatic cancer poses utmost clinical challenges and may present molecular and cellular features distinct from an early-stage cancer. Herein, we present single-cell tran- scriptome profiling of metastatic lung adenocarcinoma, the most prevalent histological lung cancer type diagnosed at stage IV in over 40% of all cases. From 208,506 cells populating the normal tissues or early to metastatic stage cancer in 44 patients, we identify a cancer cell subtype deviating from the normal differentiation trajectory and dominating the metastatic stage. In all stages, the stromal and immune cell dynamics reveal ontological and functional changes that create a pro-tumoral and immunosuppressive microenvironment. Normal resident myeloid cell populations are gradually replaced with monocyte-derived macrophages and dendritic cells, along with T-cell exhaustion. This extensive single-cell analysis enhances our understanding of molecular and cellular dynamics in metastatic lung cancer and reveals potential diagnostic and therapeutic targets in cancer-microenvironment interactions. 1 Samsung Genome Institute, Samsung Medical Center, Seoul 06351, Korea. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

TSHZ3 Deletion Causes an Autism Syndrome and Defects in Cortical Projection Neurons
Europe PMC Funders Group Author Manuscript Nat Genet. Author manuscript; available in PMC 2017 March 26. Published in final edited form as: Nat Genet. 2016 November ; 48(11): 1359–1369. doi:10.1038/ng.3681. Europe PMC Funders Author Manuscripts TSHZ3 deletion causes an autism syndrome and defects in cortical projection neurons Xavier Caubit#1, Paolo Gubellini#1, Joris Andrieux2, Pierre L. Roubertoux3, Mehdi Metwaly1, Bernard Jacq1, Ahmed Fatmi1, Laurence Had-Aissouni1, Kenneth Y. Kwan4,5, Pascal Salin1, Michèle Carlier6, Agne Liedén7, Eva Rudd7, Marwan Shinawi8, Catherine Vincent-Delorme9, Jean-Marie Cuisset10, Marie-Pierre Lemaitre10, Fatimetou Abderrehamane2, Bénédicte Duban11, Jean-François Lemaitre11, Adrian S. Woolf12, Detlef Bockenhauer13, Dany Severac14, Emeric Dubois14, Ying Zhu4, Nenad Sestan4, Alistair N. Garratt15, Lydia Kerkerian-Le Goff1, and Laurent Fasano1 1Aix Marseille Univ, CNRS, IBDM, Marseille, France 2Institut de génétique médicale, Hôpital Jeanne de Flandre, CHRU Lille, France 3Aix Marseille Univ, INSERM, GMGF, Marseille, France 4Department of Neuroscience, Yale School of Medicine, New Haven, CT, USA 5Molecular & Behavioral Neuroscience Institute (MBNI), Department of Human Genetics, University of Michigan, Ann Arbor, MI, USA 6Aix Marseille Univ, CNRS, LPC, Marseille, France Europe PMC Funders Author Manuscripts 7Karolinska University Hospital Solna, Clinical Genetics Unit, Stockholm, Sweden 8Department of Pediatrics, Division of Genetics and Genomic Medicine, Washington University School of Medicine, St. Louis, MO, USA 9Service de Génétique clinique, Hôpital Jeanne de Flandre, CHRU Lille, France 10Service de Neuropédiatrie, Hôpital Salengro, CHRU Lille, France Users may view, print, copy, and download text and data-mine the content in such documents, for the purposes of academic research, subject always to the full Conditions of use:http://www.nature.com/authors/editorial_policies/license.html#terms Correspondence should be addressed to L.F. -

The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease Jonathan Z
REVIEW pubs.acs.org/CR The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease Jonathan Z. Long* and Benjamin F. Cravatt* The Skaggs Institute for Chemical Biology and Department of Chemical Physiology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, California 92037, United States CONTENTS 2.4. Other Phospholipases 6034 1. Introduction 6023 2.4.1. LIPG (Endothelial Lipase) 6034 2. Small-Molecule Hydrolases 6023 2.4.2. PLA1A (Phosphatidylserine-Specific 2.1. Intracellular Neutral Lipases 6023 PLA1) 6035 2.1.1. LIPE (Hormone-Sensitive Lipase) 6024 2.4.3. LIPH and LIPI (Phosphatidic Acid-Specific 2.1.2. PNPLA2 (Adipose Triglyceride Lipase) 6024 PLA1R and β) 6035 2.1.3. MGLL (Monoacylglycerol Lipase) 6025 2.4.4. PLB1 (Phospholipase B) 6035 2.1.4. DAGLA and DAGLB (Diacylglycerol Lipase 2.4.5. DDHD1 and DDHD2 (DDHD Domain R and β) 6026 Containing 1 and 2) 6035 2.1.5. CES3 (Carboxylesterase 3) 6026 2.4.6. ABHD4 (Alpha/Beta Hydrolase Domain 2.1.6. AADACL1 (Arylacetamide Deacetylase-like 1) 6026 Containing 4) 6036 2.1.7. ABHD6 (Alpha/Beta Hydrolase Domain 2.5. Small-Molecule Amidases 6036 Containing 6) 6027 2.5.1. FAAH and FAAH2 (Fatty Acid Amide 2.1.8. ABHD12 (Alpha/Beta Hydrolase Domain Hydrolase and FAAH2) 6036 Containing 12) 6027 2.5.2. AFMID (Arylformamidase) 6037 2.2. Extracellular Neutral Lipases 6027 2.6. Acyl-CoA Hydrolases 6037 2.2.1. PNLIP (Pancreatic Lipase) 6028 2.6.1. FASN (Fatty Acid Synthase) 6037 2.2.2. PNLIPRP1 and PNLIPR2 (Pancreatic 2.6.2. -

Isolation and Characterization of the Prolyl Aminopeptidase Gene (Pap) from Aeromonas Sobria: Comparison with the Bacillus Coagulans Enzyme1
J. Biochem. 116, 818-825 (1994) Isolation and Characterization of the Prolyl Aminopeptidase Gene (pap) from Aeromonas sobria: Comparison with the Bacillus coagulans Enzyme1 Ana Kitazono,* Atsuko Kitano,* Daisuke Tsuru,•õ and Tadashi Yoshimoto*,2 *School of Pharmaceutical Sciences , Nagasaki University, 1-14 Bunkyo-machi, Nagasaki, Nagasaki 852; and •õ Department of Applied Microbiology, Kumamoto Institute of Technology, 4-22-1 Ikeda, Kumamoto, Kumamoto 860 Received for publication, May 16, 1994 The Aeromonas sobria pap gene encoding prolyl aminopeptidase (PAP) was cloned. It consists of 425 codons and encodes a homotetrameric enzyme of 205kDa. The purified enzyme showed an almost absolute specificity for amino-terminal proline. Proline and hydroxyproline residues from many peptide and amide substrates could be easily removed, while no activity was detected for substrates having other amino terminals. The enzyme was very similar to that from Bacillus coagulans in many aspects, such as the strong inhibition caused by PCMB and the weak or no inhibition caused by DFP and chelators, respectively. However, these enzymes show only 15% identity in their amino acid sequences. Differences were also observed in their molecular weight, stability and activity toward some peptide substrates. When aligning the deduced amino acid sequence with known sequences from other microorganisms, conserved sequences were found at the amino-terminal region; the significance of these conserved regions is discussed. Based on the results of this work, and on the studies available to date, the occurrence of at least two types of PAPs is postulated. One group would be formed by the Bacillus, Neisseria, and Lactobacillus enzymes, and the other by enzymes such as the Aeromonas PAP. -

RAMP1 and RAMP3 Differentially Control Amylin's Effects on Food
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2020 RAMP1 and RAMP3 differentially control amylin’s effects on food intake, glucose and energy balance in male and female mice Coester, Bernd Posted at the Zurich Open Repository and Archive, University of Zurich ZORA URL: https://doi.org/10.5167/uzh-191827 Dissertation Published Version Originally published at: Coester, Bernd. RAMP1 and RAMP3 differentially control amylin’s effects on food intake, glucose and energy balance in male and female mice. 2020, University of Zurich, Vetsuisse Faculty. Institut für Veterinärphysiologie der Vetsuisse-Fakultät Universität Zürich Direktor: Prof. Prof. h.c. Dr. med. vet. Max Gassmann Arbeit unter wissenschaftlicher Betreuung von Christelle Le Foll, PhD RAMP1 and RAMP3 Differentially Control Amylin’s Effects on Food Intake, Glucose and Energy Balance in Male and Female Mice Inaugural-Dissertation zur Erlangung der Doktorwürde der Vetsuisse-Fakultät Universität Zürich vorgelegt von Bernd Coester Tierarzt von Zürich, ZH genehmigt auf Antrag von Prof. Dr. med. vet. Thomas Lutz, Referent 2020 Inhaltsverzeichnis Zusammenfassung 4 Abstract 5 Introduction 6 Experimental Procedures 7 Results 9 Discussion 19 References 24 Appendix 26 3 RAMP1 und RAMP3 kontrollieren die Effekte von Amylin auf Futteraufnahme, Glukose und Energiehaushalt in männlichen und weiblichen Mäusen Bernd Coester, Sydney W Pence, Soraya Arrigoni, Christina N Boyle, Christelle Le Foll, Thomas A Lutz Amylin ist ein Peptid aus dem endokrinen Pankreas und nimmt eine Schlüsselrolle in der Kontrolle von Futteraufnahme und Energiehaushalt ein, wobei es mehrheitlich an drei Rezeptoren bindet (AMY 1-3). AMY 1-3 bestehen aus einem Calcitonin- Rezeptor (CTR) und jeweils einem rezeptor-aktivität-modifizierenden Protein (RAMP1-3). -

P-Glycoprotein, CYP3A, and Plasma Carboxylesterase Determine Brain and Blood Disposition of the Mtor Inhibitor Everolimus (Afinitor) in Mice
Published OnlineFirst April 11, 2014; DOI: 10.1158/1078-0432.CCR-13-1759 Clinical Cancer Cancer Therapy: Preclinical Research P-Glycoprotein, CYP3A, and Plasma Carboxylesterase Determine Brain and Blood Disposition of the mTOR Inhibitor Everolimus (Afinitor) in Mice Seng Chuan Tang1, Rolf W. Sparidans3, Ka Lei Cheung4, Tatsuki Fukami5, Selvi Durmus1, Els Wagenaar1, Tsuyoshi Yokoi5, Bart J.M. van Vlijmen4, Jos H. Beijnen2,3, and Alfred H. Schinkel1 Abstract Purpose: To clarify the role of ABCB1, ABCG2, and CYP3A in blood and brain exposure of everolimus using knockout mouse models. À À À À À À À À Experimental Design: We used wild-type, Abcb1a/1b / , Abcg2 / , Abcb1a/1b;Abcg2 / , and Cyp3a / mice to study everolimus oral bioavailability and brain accumulation. Results: Following everolimus administration, brain concentrations and brain-to-liver ratios were À À À À À À substantially increased in Abcb1a/1b / and Abcb1a/1b;Abcg2 / , but not Abcg2 / mice. The fraction of everolimus located in the plasma compartment was highly increased in all knockout strains. In vitro, everolimus was rapidly degraded in wild-type but not knockout plasma. Carboxylesterase 1c (Ces1c), a plasma carboxylesterase gene, was highly upregulated (80-fold) in the liver of knockout mice relative to wild-type mice, and plasma Ces1c likely protected everolimus from degradation by binding and stabilizing it. This binding was prevented by preincubation with the carboxylesterase inhibitor BNPP. In vivo knockdown experiments confirmed the involvement of Ces1c in everolimus stabilization. Everolimus also markedly inhibited the hydrolysis of irinotecan and p-nitrophenyl acetate by mouse plasma carboxylesterase À À and recombinant human CES2, respectively. -

CGRP Signaling Via CALCRL Increases Chemotherapy Resistance and Stem Cell Properties in Acute Myeloid Leukemia
International Journal of Molecular Sciences Article CGRP Signaling via CALCRL Increases Chemotherapy Resistance and Stem Cell Properties in Acute Myeloid Leukemia 1,2 1,2, 1,2, 1,2 Tobias Gluexam , Alexander M. Grandits y, Angela Schlerka y, Chi Huu Nguyen , Julia Etzler 1,2 , Thomas Finkes 1,2, Michael Fuchs 3, Christoph Scheid 3, Gerwin Heller 1,2 , Hubert Hackl 4 , Nathalie Harrer 5, Heinz Sill 6 , Elisabeth Koller 7 , Dagmar Stoiber 8,9, Wolfgang Sommergruber 10 and Rotraud Wieser 1,2,* 1 Division of Oncology, Department of Medicine I, Medical University of Vienna, Waehringer Guertel 18-20, 1090 Vienna, Austria; [email protected] (T.G.); [email protected] (A.M.G.); [email protected] (A.S.); [email protected] (C.H.N.); [email protected] (J.E.); thomas.fi[email protected] (T.F.); [email protected] (G.H.) 2 Comprehensive Cancer Center, Spitalgasse 23, 1090 Vienna, Austria 3 Department I of Internal Medicine, Center for Integrated Oncology Aachen Bonn Cologne Duesseldorf, University of Cologne, Kerpener Str. 62, 50937 Cologne, Germany; [email protected] (M.F.); [email protected] (C.S.) 4 Institute of Bioinformatics, Biocenter, Medical University of Innsbruck, Innrain 80, 6020 Innsbruck, Austria; [email protected] 5 Department for Cancer Research, Boehringer Ingelheim RCV GmbH & Co KG, Dr. Boehringer-Gasse 5-11, A-1121 Vienna, Austria; [email protected] 6 Division of Hematology, Medical University of Graz, Auenbruggerplatz -

Table SII. Significantly Differentially Expressed Mrnas of GSE23558 Data Series with the Criteria of Adjusted P<0.05 And
Table SII. Significantly differentially expressed mRNAs of GSE23558 data series with the criteria of adjusted P<0.05 and logFC>1.5. Probe ID Adjusted P-value logFC Gene symbol Gene title A_23_P157793 1.52x10-5 6.91 CA9 carbonic anhydrase 9 A_23_P161698 1.14x10-4 5.86 MMP3 matrix metallopeptidase 3 A_23_P25150 1.49x10-9 5.67 HOXC9 homeobox C9 A_23_P13094 3.26x10-4 5.56 MMP10 matrix metallopeptidase 10 A_23_P48570 2.36x10-5 5.48 DHRS2 dehydrogenase A_23_P125278 3.03x10-3 5.40 CXCL11 C-X-C motif chemokine ligand 11 A_23_P321501 1.63x10-5 5.38 DHRS2 dehydrogenase A_23_P431388 2.27x10-6 5.33 SPOCD1 SPOC domain containing 1 A_24_P20607 5.13x10-4 5.32 CXCL11 C-X-C motif chemokine ligand 11 A_24_P11061 3.70x10-3 5.30 CSAG1 chondrosarcoma associated gene 1 A_23_P87700 1.03x10-4 5.25 MFAP5 microfibrillar associated protein 5 A_23_P150979 1.81x10-2 5.25 MUCL1 mucin like 1 A_23_P1691 2.71x10-8 5.12 MMP1 matrix metallopeptidase 1 A_23_P350005 2.53x10-4 5.12 TRIML2 tripartite motif family like 2 A_24_P303091 1.23x10-3 4.99 CXCL10 C-X-C motif chemokine ligand 10 A_24_P923612 1.60x10-5 4.95 PTHLH parathyroid hormone like hormone A_23_P7313 6.03x10-5 4.94 SPP1 secreted phosphoprotein 1 A_23_P122924 2.45x10-8 4.93 INHBA inhibin A subunit A_32_P155460 6.56x10-3 4.91 PICSAR P38 inhibited cutaneous squamous cell carcinoma associated lincRNA A_24_P686965 8.75x10-7 4.82 SH2D5 SH2 domain containing 5 A_23_P105475 7.74x10-3 4.70 SLCO1B3 solute carrier organic anion transporter family member 1B3 A_24_P85099 4.82x10-5 4.67 HMGA2 high mobility group AT-hook 2 A_24_P101651 -
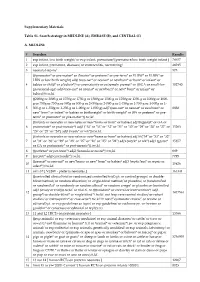
EMBASE (B), and CENTRAL (C) A. MEDLINE # Searches
Supplementary Materials Table S1. Search strategy in MEDLINE (A), EMBASE (B), and CENTRAL (C) A. MEDLINE # Searches Results 1 exp infant, low birth weight/ or exp infant, premature/ [premature/low birth weight infant ] 78657 2 exp infant, premature, diseases/ or enterocolitis, necrotizing/ 46015 3 neonatal sepsis/ 575 (((prematur* or pre-matur* or i?matur* or preterm* or pre-term* or VLBW* or ELBW* or LBW or low birth weight) adj6 (neo-nat* or neonat* or newborn* or born* or infant* or 4 babies or child* or p?ediatr*)) or prematurity or extremely premat* or ((SGA or small-for- 102740 gestational-age) adj6 (neo-nat* or neonat* or newborn* or new* born* or infant* or babies))).tw,kf. ((2000g or 2000-g or 1750g or 1750-g or 1500g or 1500-g or 1250g or 1250-g or 1000g or 1000- g or 750g or 750-g or 500g or 500-g or 2-000g or 2-000-g or 1-750g or 1-750-g or 1-500g or 1- 5 500-g or 1-250g or 1-250-g or 1-000g or 1-000-g) adj7 (neo-nat* or neonat* or newborn* or 8838 new* born* or infant* or babies or birthweight* or birth weight* or BW or preterm* or pre- term* or prematur* or pre-matur*)).tw,kf. ((infants or neonates or neo-nates or new*borns or born* or babies) adj18 (gestat* or GA or 6 postmenstr* or post-menstr*) adj3 ("34" or "33" or "32" or "31" or "30" or "29" or "28" or "27" or 15263 "26" or "25" or "24") adj3 (week* or wk*)).tw,kf. -

Supplementary Table 1 from Peeters Et Al., Mol Pharmacol, Doi: 10.1124/Mol.104.000950
Supplementary Table 1 from Peeters et al., Mol Pharmacol, doi: 10.1124/mol.104.000950 Affymetrix identified by ID SMA CFA OmniViz Gene Title Gene Symbol 100133_at yes yes yes Fyn proto-oncogene Fyn 100533_s_at yes yes yes cAMP responsive element modulator Crem 101196_at yes yes yes paired basic amino acid cleaving system 4 Pace4 101217_at yes yes yes --- --- 101426_at yes yes yes ceramide kinase Cerk 102231_at yes yes yes cAMP responsive element binding protein 3-like 1 Creb3l1 102239_at yes yes yes B-cell leukemia/lymphoma 3 Bcl3 102362_i_at yes yes yes Jun-B oncogene Junb 102363_r_at yes yes yes Jun-B oncogene Junb 102371_at yes yes yes nuclear receptor subfamily 4, group A, member 1 Nr4a1 102955_at yes yes yes nuclear factor, interleukin 3, regulated Nfil3 104008_at yes yes yes SAM pointed domain containing ets transcription factor Spdef 104410_at yes yes yes midnolin Midn 104598_at yes yes yes dual specificity phosphatase 1 Dusp1 104669_at yes yes yes interferon regulatory factor 7 Irf7 92205_at yes yes yes insulin receptor substrate 2 Irs2 92248_at yes yes yes nuclear receptor subfamily 4, group A, member 2 Nr4a2 92249_g_at yes yes yes nuclear receptor subfamily 4, group A, member 2 Nr4a2 92310_at yes yes yes polo-like kinase 2 (Drosophila) Plk2 92368_at yes yes yes receptor (calcitonin) activity modifying protein 3 Ramp3 92406_at yes yes yes CD7 antigen Cd7 1 92423_at yes yes yes par-6 (partitioning defective 6,) homolog alpha (C. elegans) Pard6a 92820_at yes yes yes ubiquitin specific protease 2 Usp2 92821_at yes yes yes ubiquitin -

A Single-Cell Transcriptome Atlas of the Mouse Glomerulus
RAPID COMMUNICATION www.jasn.org A Single-Cell Transcriptome Atlas of the Mouse Glomerulus Nikos Karaiskos,1 Mahdieh Rahmatollahi,2 Anastasiya Boltengagen,1 Haiyue Liu,1 Martin Hoehne ,2 Markus Rinschen,2,3 Bernhard Schermer,2,4,5 Thomas Benzing,2,4,5 Nikolaus Rajewsky,1 Christine Kocks ,1 Martin Kann,2 and Roman-Ulrich Müller 2,4,5 Due to the number of contributing authors, the affiliations are listed at the end of this article. ABSTRACT Background Three different cell types constitute the glomerular filter: mesangial depending on cell location relative to the cells, endothelial cells, and podocytes. However, to what extent cellular heteroge- glomerular vascular pole.3 Because BP ad- neity exists within healthy glomerular cell populations remains unknown. aptation and mechanoadaptation of glo- merular cells are key determinants of kidney Methods We used nanodroplet-based highly parallel transcriptional profiling to function and dysregulated in kidney disease, characterize the cellular content of purified wild-type mouse glomeruli. we tested whether glomerular cell type sub- Results Unsupervised clustering of nearly 13,000 single-cell transcriptomes identi- sets can be identified by single-cell RNA fied the three known glomerular cell types. We provide a comprehensive online sequencing in wild-type glomeruli. This atlas of gene expression in glomerular cells that can be queried and visualized using technique allows for high-throughput tran- an interactive and freely available database. Novel marker genes for all glomerular scriptome profiling of individual cells and is cell types were identified and supported by immunohistochemistry images particularly suitable for identifying novel obtained from the Human Protein Atlas.