A Middle-Aged Enzyme Still in Its Prime: Recent Advances in the Field of Cutinases
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Kramers' Theory and the Dependence of Enzyme Dynamics on Trehalose
catalysts Review Kramers’ Theory and the Dependence of Enzyme Dynamics on Trehalose-Mediated Viscosity José G. Sampedro 1,* , Miguel A. Rivera-Moran 1 and Salvador Uribe-Carvajal 2 1 Instituto de Física, Universidad Autónoma de San Luis Potosí, Manuel Nava 6, Zona Universitaria, San Luis Potosí C.P. 78290, Mexico; [email protected] 2 Instituto de Fisiología Celular, Universidad Nacional Autónoma de México, Ciudad de México C.P. 04510, Mexico; [email protected] * Correspondence: [email protected]fisica.uaslp.mx; Tel.: +52-(444)-8262-3200 (ext. 5715) Received: 29 April 2020; Accepted: 29 May 2020; Published: 11 June 2020 Abstract: The disaccharide trehalose is accumulated in the cytoplasm of some organisms in response to harsh environmental conditions. Trehalose biosynthesis and accumulation are important for the survival of such organisms by protecting the structure and function of proteins and membranes. Trehalose affects the dynamics of proteins and water molecules in the bulk and the protein hydration shell. Enzyme catalysis and other processes dependent on protein dynamics are affected by the viscosity generated by trehalose, as described by the Kramers’ theory of rate reactions. Enzyme/protein stabilization by trehalose against thermal inactivation/unfolding is also explained by the viscosity mediated hindering of the thermally generated structural dynamics, as described by Kramers’ theory. The analysis of the relationship of viscosity–protein dynamics, and its effects on enzyme/protein function and other processes (thermal inactivation and unfolding/folding), is the focus of the present work regarding the disaccharide trehalose as the viscosity generating solute. Finally, trehalose is widely used (alone or in combination with other compounds) in the stabilization of enzymes in the laboratory and in biotechnological applications; hence, considering the effect of viscosity on catalysis and stability of enzymes may help to improve the results of trehalose in its diverse uses/applications. -

Cutinase Structure, Function and Biocatalytic Applications
EJB Electronic Journal of Biotechnology ISSN: 0717-345 Vol.1 No.3, Issue of August 15, 1998. © 1998 by Universidad Católica de Valparaíso –- Chile Invited review paper/ Received 20 October, 1998. REVIEW ARTICLE Cutinase structure, function and biocatalytic applications Cristina M. L. Carvalho Centro de Engenharia Biológica e Química Instituto Superior Técnico 1000 Lisboa – Portugal Maria Raquel Aires-Barros Centro de Engenharia Biológica e Química Instituto Superior Técnico 1000 Lisboa – Portugal 1 Joaquim M. S. Cabral Centro de Engenharia Biológica e Química Instituto Superior Técnico 1000 Lisboa – Portugal Tel: + 351.1.8419065 Fax: + 351.1.8419062 E-mail: [email protected] http://www.ist.utl.pt/ This review analyses the role of cutinases in nature and Some microorganisms, including plant pathogens, have their potential biotechnological applications. The been shown to live on cutin as their sole carbon source cloning and expression of a fungal cutinase from (Purdy and Kolattukudy, 1975) and the production of Fusarium solani f. pisi, in Escherichia coli and extracellular cutinolytic enzymes has been presented (Purdy Saccharomyces cerevisiae hosts are described. The three and Kolattukudy, 1975; Lin and Kolattukudy, 1980). dimensional structure of this cutinase is also analysed Cutinases have been purified and characterized from and its function as a lipase discussed and compared with several different sources, mainly fungi (Purdy and other lipases. The biocatalytic applications of cutinase Kolattukudy, 1975 ; Lin and Kolattukudy, 1980; Petersen et are described taking into account the preparation of al., 1997; Dantzig et al., 1986; Murphy et al., 1996) and different cutinase forms and the media where the pollen (Petersen et al., 1997; Sebastian et al., 1987). -

Cutinases from Mycobacterium Tuberculosis
Identification of Residues Involved in Substrate Specificity and Cytotoxicity of Two Closely Related Cutinases from Mycobacterium tuberculosis Luc Dedieu, Carole Serveau-Avesque, Ste´phane Canaan* CNRS - Aix-Marseille Universite´ - Enzymologie Interfaciale et Physiologie de la Lipolyse - UMR 7282, Marseille, France Abstract The enzymes belonging to the cutinase family are serine enzymes active on a large panel of substrates such as cutin, triacylglycerols, and phospholipids. In the M. tuberculosis H37Rv genome, seven genes coding for cutinase-like proteins have been identified with strong immunogenic properties suggesting a potential role as vaccine candidates. Two of these enzymes which are secreted and highly homologous, possess distinct substrates specificities. Cfp21 is a lipase and Cut4 is a phospholipase A2, which has cytotoxic effects on macrophages. Structural overlay of their three-dimensional models allowed us to identify three areas involved in the substrate binding process and to shed light on this substrate specificity. By site-directed mutagenesis, residues present in these Cfp21 areas were replaced by residues occurring in Cut4 at the same location. Three mutants acquired phospholipase A1 and A2 activities and the lipase activities of two mutants were 3 and 15 fold greater than the Cfp21 wild type enzyme. In addition, contrary to mutants with enhanced lipase activity, mutants that acquired phospholipase B activities induced macrophage lysis as efficiently as Cut4 which emphasizes the relationship between apparent phospholipase A2 activity and cytotoxicity. Modification of areas involved in substrate specificity, generate recombinant enzymes with higher activity, which may be more immunogenic than the wild type enzymes and could therefore constitute promising candidates for antituberculous vaccine production. -

Purification and Characterization of Cutinase from Venturia Inaequalis
Physiology and Biochemistry Purification and Characterization of Cutinase from Venturia inaequalis Wolfram K611er and Diana M. Parker Assistant professor and research assistant, Department of Plant Pathology, Cornell University, New York State Agricultural Experiment Station, Geneva 14456. We thank Professor A. L. Jones for the isolates of Venturia inaequalis, Mr. Robert W. Ennis, Jr. for his help in cutin preparation, and Comstock Foods, Alton, NY, for the apple peels. Accepted for publication 16 August 1988. ABSTRACT K61ler, W., and Parker, D. M. 1989. Purification and characterization of cutinase from Venturia inaequalis. Phytopathology 79:278-283. Venturia inaequalis was grown in a culture medium containing purified cutinase from V. inaequalisis optimal at a pH of 6 and thus different from apple cutin as the sole carbon source. After 8 wk of growth an esterase was the alkaline pH-optimum reported for other purified cutinases. The isolated from the culture fluid and purified to apparent homogeneity. The hydrolysis of the model esterp-nitrophenyl butyrate was less affected by the enzyme hydrolyzed tritiated cutin and thus was identified as cutinase. The pH. The esterase activity was strongly inhibited by diisopropyl purified cutinase is a glycoprotein with a molecular mass of 21-23 kg/ mol, fluorophosphate, and the phosphorylation of one serine was sufficient for as determined by various procedures. Remarkable structural features are a complete inhibition. Thus, cutinase from V. inaequalis belongs to the class high content of glycine, a high content of nonpolar amino acids, two of serine hydrolases, a characterisitic shared with other fungal cutinases. disulfide bridges, and a high degree of hydrophobicity. -

Letters to Nature
letters to nature Received 7 July; accepted 21 September 1998. 26. Tronrud, D. E. Conjugate-direction minimization: an improved method for the re®nement of macromolecules. Acta Crystallogr. A 48, 912±916 (1992). 1. Dalbey, R. E., Lively, M. O., Bron, S. & van Dijl, J. M. The chemistry and enzymology of the type 1 27. Wolfe, P. B., Wickner, W. & Goodman, J. M. Sequence of the leader peptidase gene of Escherichia coli signal peptidases. Protein Sci. 6, 1129±1138 (1997). and the orientation of leader peptidase in the bacterial envelope. J. Biol. Chem. 258, 12073±12080 2. Kuo, D. W. et al. Escherichia coli leader peptidase: production of an active form lacking a requirement (1983). for detergent and development of peptide substrates. Arch. Biochem. Biophys. 303, 274±280 (1993). 28. Kraulis, P.G. Molscript: a program to produce both detailed and schematic plots of protein structures. 3. Tschantz, W. R. et al. Characterization of a soluble, catalytically active form of Escherichia coli leader J. Appl. Crystallogr. 24, 946±950 (1991). peptidase: requirement of detergent or phospholipid for optimal activity. Biochemistry 34, 3935±3941 29. Nicholls, A., Sharp, K. A. & Honig, B. Protein folding and association: insights from the interfacial and (1995). the thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet. 11, 281±296 (1991). 4. Allsop, A. E. et al.inAnti-Infectives, Recent Advances in Chemistry and Structure-Activity Relationships 30. Meritt, E. A. & Bacon, D. J. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277, 505± (eds Bently, P. H. & O'Hanlon, P. J.) 61±72 (R. Soc. Chem., Cambridge, 1997). -

The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease Jonathan Z
REVIEW pubs.acs.org/CR The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease Jonathan Z. Long* and Benjamin F. Cravatt* The Skaggs Institute for Chemical Biology and Department of Chemical Physiology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, California 92037, United States CONTENTS 2.4. Other Phospholipases 6034 1. Introduction 6023 2.4.1. LIPG (Endothelial Lipase) 6034 2. Small-Molecule Hydrolases 6023 2.4.2. PLA1A (Phosphatidylserine-Specific 2.1. Intracellular Neutral Lipases 6023 PLA1) 6035 2.1.1. LIPE (Hormone-Sensitive Lipase) 6024 2.4.3. LIPH and LIPI (Phosphatidic Acid-Specific 2.1.2. PNPLA2 (Adipose Triglyceride Lipase) 6024 PLA1R and β) 6035 2.1.3. MGLL (Monoacylglycerol Lipase) 6025 2.4.4. PLB1 (Phospholipase B) 6035 2.1.4. DAGLA and DAGLB (Diacylglycerol Lipase 2.4.5. DDHD1 and DDHD2 (DDHD Domain R and β) 6026 Containing 1 and 2) 6035 2.1.5. CES3 (Carboxylesterase 3) 6026 2.4.6. ABHD4 (Alpha/Beta Hydrolase Domain 2.1.6. AADACL1 (Arylacetamide Deacetylase-like 1) 6026 Containing 4) 6036 2.1.7. ABHD6 (Alpha/Beta Hydrolase Domain 2.5. Small-Molecule Amidases 6036 Containing 6) 6027 2.5.1. FAAH and FAAH2 (Fatty Acid Amide 2.1.8. ABHD12 (Alpha/Beta Hydrolase Domain Hydrolase and FAAH2) 6036 Containing 12) 6027 2.5.2. AFMID (Arylformamidase) 6037 2.2. Extracellular Neutral Lipases 6027 2.6. Acyl-CoA Hydrolases 6037 2.2.1. PNLIP (Pancreatic Lipase) 6028 2.6.1. FASN (Fatty Acid Synthase) 6037 2.2.2. PNLIPRP1 and PNLIPR2 (Pancreatic 2.6.2. -

Isolation and Characterization of the Prolyl Aminopeptidase Gene (Pap) from Aeromonas Sobria: Comparison with the Bacillus Coagulans Enzyme1
J. Biochem. 116, 818-825 (1994) Isolation and Characterization of the Prolyl Aminopeptidase Gene (pap) from Aeromonas sobria: Comparison with the Bacillus coagulans Enzyme1 Ana Kitazono,* Atsuko Kitano,* Daisuke Tsuru,•õ and Tadashi Yoshimoto*,2 *School of Pharmaceutical Sciences , Nagasaki University, 1-14 Bunkyo-machi, Nagasaki, Nagasaki 852; and •õ Department of Applied Microbiology, Kumamoto Institute of Technology, 4-22-1 Ikeda, Kumamoto, Kumamoto 860 Received for publication, May 16, 1994 The Aeromonas sobria pap gene encoding prolyl aminopeptidase (PAP) was cloned. It consists of 425 codons and encodes a homotetrameric enzyme of 205kDa. The purified enzyme showed an almost absolute specificity for amino-terminal proline. Proline and hydroxyproline residues from many peptide and amide substrates could be easily removed, while no activity was detected for substrates having other amino terminals. The enzyme was very similar to that from Bacillus coagulans in many aspects, such as the strong inhibition caused by PCMB and the weak or no inhibition caused by DFP and chelators, respectively. However, these enzymes show only 15% identity in their amino acid sequences. Differences were also observed in their molecular weight, stability and activity toward some peptide substrates. When aligning the deduced amino acid sequence with known sequences from other microorganisms, conserved sequences were found at the amino-terminal region; the significance of these conserved regions is discussed. Based on the results of this work, and on the studies available to date, the occurrence of at least two types of PAPs is postulated. One group would be formed by the Bacillus, Neisseria, and Lactobacillus enzymes, and the other by enzymes such as the Aeromonas PAP. -

P-Glycoprotein, CYP3A, and Plasma Carboxylesterase Determine Brain and Blood Disposition of the Mtor Inhibitor Everolimus (Afinitor) in Mice
Published OnlineFirst April 11, 2014; DOI: 10.1158/1078-0432.CCR-13-1759 Clinical Cancer Cancer Therapy: Preclinical Research P-Glycoprotein, CYP3A, and Plasma Carboxylesterase Determine Brain and Blood Disposition of the mTOR Inhibitor Everolimus (Afinitor) in Mice Seng Chuan Tang1, Rolf W. Sparidans3, Ka Lei Cheung4, Tatsuki Fukami5, Selvi Durmus1, Els Wagenaar1, Tsuyoshi Yokoi5, Bart J.M. van Vlijmen4, Jos H. Beijnen2,3, and Alfred H. Schinkel1 Abstract Purpose: To clarify the role of ABCB1, ABCG2, and CYP3A in blood and brain exposure of everolimus using knockout mouse models. À À À À À À À À Experimental Design: We used wild-type, Abcb1a/1b / , Abcg2 / , Abcb1a/1b;Abcg2 / , and Cyp3a / mice to study everolimus oral bioavailability and brain accumulation. Results: Following everolimus administration, brain concentrations and brain-to-liver ratios were À À À À À À substantially increased in Abcb1a/1b / and Abcb1a/1b;Abcg2 / , but not Abcg2 / mice. The fraction of everolimus located in the plasma compartment was highly increased in all knockout strains. In vitro, everolimus was rapidly degraded in wild-type but not knockout plasma. Carboxylesterase 1c (Ces1c), a plasma carboxylesterase gene, was highly upregulated (80-fold) in the liver of knockout mice relative to wild-type mice, and plasma Ces1c likely protected everolimus from degradation by binding and stabilizing it. This binding was prevented by preincubation with the carboxylesterase inhibitor BNPP. In vivo knockdown experiments confirmed the involvement of Ces1c in everolimus stabilization. Everolimus also markedly inhibited the hydrolysis of irinotecan and p-nitrophenyl acetate by mouse plasma carboxylesterase À À and recombinant human CES2, respectively. -

Mycobacterial Lipolytic Enzymes: a Gold Mine for Tuberculosis Research Luc Dedieu, C
Mycobacterial lipolytic enzymes: A gold mine for tuberculosis research Luc Dedieu, C. Serveau-Avesque, L. Kremer, Stéphane Canaan To cite this version: Luc Dedieu, C. Serveau-Avesque, L. Kremer, Stéphane Canaan. Mycobacterial lipolytic en- zymes: A gold mine for tuberculosis research. Biochimie, Elsevier, 2013, 95 (1), pp.66-73. 10.1016/j.biochi.2012.07.008. hal-02646757 HAL Id: hal-02646757 https://hal.inrae.fr/hal-02646757 Submitted on 29 May 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - NonCommercial - NoDerivatives| 4.0 International License Version définitive du manuscrit publiée dans / Final version of the manuscript published in : Biochimie (2013), Vol. 95, p. 66-73, DOI: 10.1016/j.biochi.2012.07.008 Journal homepage : www.elsevier.com/locate/biochi ipt cr nus a Review m Mycobacterial lipolytic enzymes: A gold mine for tuberculosis research a a b,c a,* / Author L. Dedieu , C. Serveau-Avesque , L. Kremer , S. Canaan r a u CNRS UMR7282-Aix-Marseille Université e Enzymologie Interfaciale -

WO 2013/016115 Al 31 January 2013 (31.01.2013) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2013/016115 Al 31 January 2013 (31.01.2013) P O P C T (51) International Patent Classification: (81) Designated States (unless otherwise indicated, for every CUP 19/14 (2006.01) kind of national protection available): AE, AG, AL, AM, AO, AT, AU, AZ, BA, BB, BG, BH, BR, BW, BY, BZ, (21) International Application Number: CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, DO, PCT/US20 12/047326 DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, (22) International Filing Date: HR, HU, ID, IL, IN, IS, JP, KE, KG, KM, KN, KP, KR, 19 July 2012 (19.07.2012) KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, (25) Filing Language: English OM, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SC, SD, (26) Publication Language: English SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (30) Priority Data: 61/5 10,637 22 July 201 1 (22.07.201 1) US (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (71) Applicant (for all designated States except US) : NO- GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, SZ, TZ, VOZYMES NORTH AMERICA, INC. -
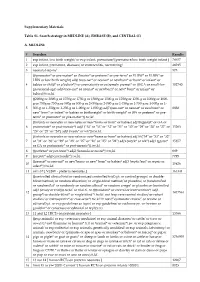
EMBASE (B), and CENTRAL (C) A. MEDLINE # Searches
Supplementary Materials Table S1. Search strategy in MEDLINE (A), EMBASE (B), and CENTRAL (C) A. MEDLINE # Searches Results 1 exp infant, low birth weight/ or exp infant, premature/ [premature/low birth weight infant ] 78657 2 exp infant, premature, diseases/ or enterocolitis, necrotizing/ 46015 3 neonatal sepsis/ 575 (((prematur* or pre-matur* or i?matur* or preterm* or pre-term* or VLBW* or ELBW* or LBW or low birth weight) adj6 (neo-nat* or neonat* or newborn* or born* or infant* or 4 babies or child* or p?ediatr*)) or prematurity or extremely premat* or ((SGA or small-for- 102740 gestational-age) adj6 (neo-nat* or neonat* or newborn* or new* born* or infant* or babies))).tw,kf. ((2000g or 2000-g or 1750g or 1750-g or 1500g or 1500-g or 1250g or 1250-g or 1000g or 1000- g or 750g or 750-g or 500g or 500-g or 2-000g or 2-000-g or 1-750g or 1-750-g or 1-500g or 1- 5 500-g or 1-250g or 1-250-g or 1-000g or 1-000-g) adj7 (neo-nat* or neonat* or newborn* or 8838 new* born* or infant* or babies or birthweight* or birth weight* or BW or preterm* or pre- term* or prematur* or pre-matur*)).tw,kf. ((infants or neonates or neo-nates or new*borns or born* or babies) adj18 (gestat* or GA or 6 postmenstr* or post-menstr*) adj3 ("34" or "33" or "32" or "31" or "30" or "29" or "28" or "27" or 15263 "26" or "25" or "24") adj3 (week* or wk*)).tw,kf. -

The Role of the B-Type Phospholipases in S. Cerevisiae: Function, Regulation, and Physiological Relevance in Lipid Homeostasis Beth a Surlow
Duquesne University Duquesne Scholarship Collection Electronic Theses and Dissertations Summer 2014 The Role of the B-Type Phospholipases in S. cerevisiae: Function, Regulation, and Physiological Relevance in Lipid Homeostasis Beth A Surlow Follow this and additional works at: https://dsc.duq.edu/etd Recommended Citation Surlow, B. (2014). The Role of the B-Type Phospholipases in S. cerevisiae: Function, Regulation, and Physiological Relevance in Lipid Homeostasis (Doctoral dissertation, Duquesne University). Retrieved from https://dsc.duq.edu/etd/1255 This Immediate Access is brought to you for free and open access by Duquesne Scholarship Collection. It has been accepted for inclusion in Electronic Theses and Dissertations by an authorized administrator of Duquesne Scholarship Collection. For more information, please contact [email protected]. THE ROLE OF B-TYPE PHOSPHOLIPASES IN S. CEREVISIAE: FUNCTION, REGULATION, AND PHYSIOLOGICAL RELEVANCE IN LIPID HOMEOSTASIS A Dissertation Submitted to the Bayer School of Natural and Environmental Sciences Duquesne University In partial fulfillment of the requirements for the degree of Doctor of Philosophy By Beth A. Surlow August 2014 Copyright by Beth A. Surlow 2014 THE ROLE OF B-TYPE PHOSPHOLIPASES IN S. CEREVISIAE: FUNCTION, REGULATION, AND PHYSIOLOGICAL RELEVANCE IN LIPID HOMEOSTASIS By Beth A. Surlow Approved June 5, 2014 ________________________________ ________________________________ Dr. Jana Patton-Vogt Dr. Philip Auron Associate Professor of Biological Professor of Biological Sciences Sciences (Committee Member) (Committee Chair) ________________________________ ________________________________ Dr. Joseph McCormick Dr. Jeffrey Brodsky Chair and Associate Professor of Professor of Biological Sciences Biological Sciences University of Pittsburgh (Committee Member) (Committee Member) ________________________________ Dr. Philip Reeder Dean, Bayer School of Natural and Environmental Science iii ABSTRACT THE ROLE OF B-TYPE PHOSPHOLIPASES IN S.