DNA Sequence, June 2008; 19(3): 246–255
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

ABSTRACT LUCKENBACH, JOHN ADAM. Breeding Biotechnology
ABSTRACT LUCKENBACH, JOHN ADAM. Breeding Biotechnology, Sex Determination, and Growth in Southern flounder, Paralichthys lethostigma. (Under the direction of Dr. John Godwin and Dr. Russell J. Borski). Southern flounder (Paralichthys lethostigma) support valuable, but declining US fisheries. This species is therefore a strong candidate for aquaculture to mitigate fishing impacts and stabilize seafood supply. Because female flounder reach substantially larger sizes than males, all-female culture is desirable for commercial aquaculture. Hence, a thorough understanding of sexual development, its timing and regulation by temperature is essential for optimization of flounder aquaculture. To better understand ovarian and testicular development in southern flounder, structural and cellular sex-distinguishing markers were studied using histological methods. We found that histologically discernible sex differentiation occurs in southern flounder at ~75-120 mm TL and that early differentiation is considerably delayed relative to its Japanese congener, P. olivaceus. High (28°C) and low (18°C) water temperatures, produced a higher proportion of males (96% and 78% males, respectively). The sex ratio at a mid-range (23°C) temperature was not different from 1:1. This suggests that southern flounder possess a temperature sensitive mechanism of sex determination. Growth was also affected by temperature with the temperature that maximized females inducing better growth. Aromatase cytochrome P450 (P450arom) is responsible for estrogen biosynthesis and plays a critical role in ovarian differentiation. We cloned ovarian P450arom and developed a qRT-PCR for assessment of early sex differentiation. The deduced amino acid sequence for southern flounder P450arom is very similar to P450arom in other teleosts. Comparison of P450arom intron sequences of southern flounder within and between different populations revealed substantial inter-individual variation that may affect sex determination responses. -

Disease List for Aquaculture Health Certificate
Quarantine Standard for Designated Species of Imported/Exported Aquatic Animals [Attached Table] 4. Listed Diseases & Quarantine Standard for Designated Species Listed disease designated species standard Common name Disease Pathogen 1. Epizootic haematopoietic Epizootic Perca fluviatilis Redfin perch necrosis(EHN) haematopoietic Oncorhynchus mykiss Rainbow trout necrosis virus(EHNV) Macquaria australasica Macquarie perch Bidyanus bidyanus Silver perch Gambusia affinis Mosquito fish Galaxias olidus Mountain galaxias Negative Maccullochella peelii Murray cod Salmo salar Atlantic salmon Ameirus melas Black bullhead Esox lucius Pike 2. Spring viraemia of Spring viraemia of Cyprinus carpio Common carp carp, (SVC) carp virus(SVCV) Grass carp, Ctenopharyngodon idella white amur Hypophthalmichthys molitrix Silver carp Hypophthalmichthys nobilis Bighead carp Carassius carassius Crucian carp Carassius auratus Goldfish Tinca tinca Tench Sheatfish, Silurus glanis European catfish, wels Negative Leuciscus idus Orfe Rutilus rutilus Roach Danio rerio Zebrafish Esox lucius Northern pike Poecilia reticulata Guppy Lepomis gibbosus Pumpkinseed Oncorhynchus mykiss Rainbow trout Abramis brama Freshwater bream Notemigonus cysoleucas Golden shiner 3.Viral haemorrhagic Viral haemorrhagic Oncorhynchus spp. Pacific salmon septicaemia(VHS) septicaemia Oncorhynchus mykiss Rainbow trout virus(VHSV) Gadus macrocephalus Pacific cod Aulorhynchus flavidus Tubesnout Cymatogaster aggregata Shiner perch Ammodytes hexapterus Pacific sandlance Merluccius productus Pacific -
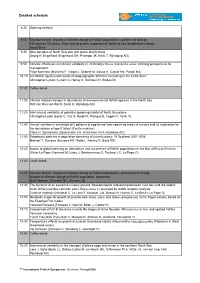
Schedule Onlinepdf
Detailed schedule 8:30 Opening session 9:00 Keynote lecture: Impacts of climate change on flatfish populations - patterns of change 100 days to 100 years: Short and long-term responses of flatfish to sea temperature change David Sims 9:30 Nine decades of North Sea sole and plaice distributions Georg H. Engelhard (Engelhard GH, Pinnegar JK, Kell LT, Rijnsdorp AD) 9:50 Climatic effects on recruitment variability in Platichthys flesus and Solea solea: defining perspectives for management. Filipe Martinho (Martinho F, Viegas I, Dolbeth M, Sousa H, Cabral HN, Pardal MA) 10:10 Are flatfish species with southern biogeographic affinities increasing in the Celtic Sea? Christopher Lynam (Lynam C, Harlay X, Gerritsen H, Stokes D) 10:30 Coffee break 11:00 Climate related changes in abundance of non-commercial flatfish species in the North Sea Ralf van Hal (van Hal R, Smits K, Rijnsdorp AD) 11:20 Inter-annual variability of potential spawning habitat of North Sea plaice Christophe Loots (Loots C, Vaz S, Koubii P, Planque B, Coppin F, Verin Y) 11:40 Annual variation in simulated drift patterns of egg/larvae from spawning areas to nursery and its implication for the abundance of age-0 turbot (Psetta maxima) Claus R. Sparrevohn (Sparrevohn CR, Hinrichsen H-H, Rijnsdorp AD) 12:00 Broadscale patterns in population dynamics of juvenile plaice: W Scotland 2001-2008 Michael T. Burrows (Burrows MT, Robb L, Harvey R, Batty RS) 12:20 Impact of global warming on abundance and occurrence of flatfish populations in the Bay of Biscay (France) Olivier Le Pape (Hermant -

Pleuronectidae
FAMILY Pleuronectidae Rafinesque, 1815 - righteye flounders [=Heterosomes, Pleronetti, Pleuronectia, Diplochiria, Poissons plats, Leptosomata, Diprosopa, Asymmetrici, Platessoideae, Hippoglossoidinae, Psettichthyini, Isopsettini] Notes: Hétérosomes Duméril, 1805:132 [ref. 1151] (family) ? Pleuronectes [latinized to Heterosomi by Jarocki 1822:133, 284 [ref. 4984]; no stem of the type genus, not available, Article 11.7.1.1] Pleronetti Rafinesque, 1810b:14 [ref. 3595] (ordine) ? Pleuronectes [published not in latinized form before 1900; not available, Article 11.7.2] Pleuronectia Rafinesque, 1815:83 [ref. 3584] (family) Pleuronectes [senior objective synonym of Platessoideae Richardson, 1836; family name sometimes seen as Pleuronectiidae] Diplochiria Rafinesque, 1815:83 [ref. 3584] (subfamily) ? Pleuronectes [no stem of the type genus, not available, Article 11.7.1.1] Poissons plats Cuvier, 1816:218 [ref. 993] (family) Pleuronectes [no stem of the type genus, not available, Article 11.7.1.1] Leptosomata Goldfuss, 1820:VIII, 72 [ref. 1829] (family) ? Pleuronectes [no stem of the type genus, not available, Article 11.7.1.1] Diprosopa Latreille, 1825:126 [ref. 31889] (family) Platessa [no stem of the type genus, not available, Article 11.7.1.1] Asymmetrici Minding, 1832:VI, 89 [ref. 3022] (family) ? Pleuronectes [no stem of the type genus, not available, Article 11.7.1.1] Platessoideae Richardson, 1836:255 [ref. 3731] (family) Platessa [junior objective synonym of Pleuronectia Rafinesque, 1815, invalid, Article 61.3.2 Hippoglossoidinae Cooper & Chapleau, 1998:696, 706 [ref. 26711] (subfamily) Hippoglossoides Psettichthyini Cooper & Chapleau, 1998:708 [ref. 26711] (tribe) Psettichthys Isopsettini Cooper & Chapleau, 1998:709 [ref. 26711] (tribe) Isopsetta SUBFAMILY Atheresthinae Vinnikov et al., 2018 - righteye flounders GENUS Atheresthes Jordan & Gilbert, 1880 - righteye flounders [=Atheresthes Jordan [D. -

Consequences of Hatchery Stocking
bioRxiv preprint doi: https://doi.org/10.1101/828798; this version posted November 2, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Consequences of hatchery stocking: 2 Lessons from Japan 3 4 Sort title: Japan hatchery stocking 5 6 Shuichi Kitada 7 8 1Tokyo University of Marine Science and Technology, Tokyo 108-8477, Japan. 9 E-mail: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/828798; this version posted November 2, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 10 Abstract 11 More than 26 billion juveniles of 180 marine species are released annually into the wild in 12 over 20 countries, but the usefulness remains unclear. Here, I review the effects of stocking by 13 Japanese marine and salmon stock-enhancement programmes, and evaluate the efficacy 14 through a meta-analysis, with new cases added to those previously considered. The posterior 15 mean recapture rates (±SD) was 8.2 ± 4.7%; mean economic efficiency was 2.7 ± 4.5, with 16 many cases having values of 1–2, excluding consideration for personnel costs and negative 17 impacts on wild populations. At the macro-scale, the proportion of released seeds to total 18 catch was 76 ± 20% for Japanese scallop, 28 ± 10% for abalone, 20 ± 5% for swimming crab, 19 13 ± 5% for kuruma prawn, 11 ± 4% for Japanese flounder and 7 ± 2% for red sea bream. -

Session Summaries-2008
Session Summaries-2008 SUMMARY OF SCIENTIFIC SESSIONS AND WORKSHOPS Science Board Symposium (S1) Beyond observations to achieving understanding and forecasting in a changing North Pacific: Forward to the FUTURE Co-Convenors: John E. Stein (SB), Michael J. Dagg (BIO), Gordon H. Kruse (FIS), Glen S. Jamieson (MEQ), Hiroya Sugisaki (MONITOR), Michael G. Foreman (POC), Bernard A. Megrey (TCODE), Harold P. Batchelder (CCCC), Michio J. Kishi (CCCC), Fangli Qiao (China), Sinjae Yoo (Korea) and Mikhail Stepanenko (Russia) Background FUTURE (Forecasting and Understanding Trends, Uncertainty and Responses of North Pacific Marine Ecosystems), the new Science Program undertaken by PICES member countries, has the broad goals of: (1) understanding the responses of marine ecosystems in the North Pacific to climate change and human activities at basin-wide and regional scales; (2) providing forecasts of what might be expected based on a current understanding of how nature works; and (3) communicating this information effectively to its members and to society in general. Past advances in understanding marine ecosystems in the North Pacific have been largely based either on the direct analysis of observations, or on the development of conceptual and numerical models that help to describe the processes underlying the observations. Though these activities will continue to play an important role in FUTURE, the provision of forecasts and estimates of their associated uncertainties necessitates moving beyond observationally based understanding, so that ecosystem responses to natural and anthropogenic changes can be anticipated and communicated effectively to society. Presentations were invited to address the goals of FUTURE and the three key research questions that it identifies: 1. -

Shellfish Diseases and Their Management in Commercial Recirculating Systems
Shellfish Diseases and Their Management in Commercial Recirculating Systems Ralph Elston AquaTechnics & Pacific Shellfish Institute PO Box 687 Carlsborg, WA 98324 Introduction Intensive culture of early life stages of bivalve shellfish culture has been practiced since at least the late 1950’s on an experimental basis. Production scale culture emerged in the 1970’s and today, hathcheries and nurseries produce large numbers of a variety of species of oysters, clams and scallops. The early life stage systems may be entirely or partially recirculating or static. Management of infectious diseases in these systems has been a challenge since their inception and effective health management is a requisite to successful culture. The diseases which affect early life stage shellfish in intensive production systems and the principles and practice of health management are the subject of this presentation. Shellfish Diseases and Management Diseases of bivalve shellfish affecting those reared or harvested from extensive culture primarily consist of parasitic infections and generally comprise the reportable or certifiable diseases. Due to the extensive nature of such culture, intervention options or disease control are limited. In contrast, infectious diseases known from early life stages in intensive culture systems tend to be opportunistic in nature and offer substantial opportunity for management due to the control that can be exerted at key points in the systems. In marine shellfish hatcheries, infectious organisms can enter the system from three sources: brood stock, seawater source and algal food source. Once an organism is established in the system, it may persist without further introduction. Bacterial infections are the most common opportunistic infection in shellfish hatcheries. -

Locus Number Estimation of MHC Class II B in Stone Flounder and Japanese Flounder
Int. J. Mol. Sci. 2015, 16, 6000-6017; doi:10.3390/ijms16036000 OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Article Locus Number Estimation of MHC Class II B in Stone Flounder and Japanese Flounder Jiajun Jiang †, Chunmei Li †, Quanqi Zhang and Xubo Wang * Key Laboratory of Marine Genetics and Breeding (Ocean University of China), Ministry of Education, Qingdao 266003, China; E-Mails: [email protected] (J.J.); [email protected] (C.L.); [email protected] (Q.Z.) † These authors contributed equally to this work. * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel./Fax: +86-532-8203-1986. Academic Editor: Ritva Tikkanen Received: 2 December 2014 / Accepted: 25 December 2014 / Published: 13 March 2015 Abstract: Members of major histocompatibility complex (MHC) family are important in immune systems. Great efforts have been made to reveal their complicated gene structures. But many existing studies focus on partial sequences of MHC genes. In this study, by gene cloning and sequencing, we identified cDNA sequences and DNA sequences of the MHC class II B in two flatfishes, stone flounder (Kareius bicoloratus) and homozygous diploid Japanese flounder (Paralichthys olivaceus). Eleven cDNA sequences were acquired from eight stone flounder individuals, and most of the polymorphic sites distributed in exons 2 and 3. Twenty-eight alleles were identified from the DNA fragments in these eight individuals. It could be deduced from their Bayesian inference phylogenetic tree that at least four loci of MHC class II B exist in stone flounder. The detailed whole-length DNA sequences in one individual were analyzed, revealing that the intron length varied among different loci. -
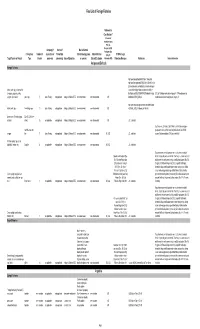
2018 Final LOFF W/ Ref and Detailed Info
Final List of Foreign Fisheries Rationale for Classification ** (Presence of mortality or injury (P/A), Co- Occurrence (C/O), Company (if Source of Marine Mammal Analogous Gear Fishery/Gear Number of aquaculture or Product (for Interactions (by group Marine Mammal (A/G), No RFMO or Legal Target Species or Product Type Vessels processor) processing) Area of Operation or species) Bycatch Estimates Information (N/I)) Protection Measures References Detailed Information Antigua and Barbuda Exempt Fisheries http://www.fao.org/fi/oldsite/FCP/en/ATG/body.htm http://www.fao.org/docrep/006/y5402e/y5402e06.htm,ht tp://www.tradeboss.com/default.cgi/action/viewcompan lobster, rock, spiny, demersal fish ies/searchterm/spiny+lobster/searchtermcondition/1/ , (snappers, groupers, grunts, ftp://ftp.fao.org/fi/DOCUMENT/IPOAS/national/Antigua U.S. LoF Caribbean spiny lobster trap/ pot >197 None documented, surgeonfish), flounder pots, traps 74 Lewis Fishing not applicable Antigua & Barbuda EEZ none documented none documented A/G AndBarbuda/NPOA_IUU.pdf Caribbean mixed species trap/pot are category III http://www.nmfs.noaa.gov/pr/interactions/fisheries/tabl lobster, rock, spiny free diving, loops 19 Lewis Fishing not applicable Antigua & Barbuda EEZ none documented none documented A/G e2/Atlantic_GOM_Caribbean_shellfish.html Queen conch (Strombus gigas), Dive (SCUBA & free molluscs diving) 25 not applicable not applicable Antigua & Barbuda EEZ none documented none documented A/G U.S. trade data Southeastern U.S. Atlantic, Gulf of Mexico, and Caribbean snapper- handline, hook and grouper and other reef fish bottom longline/hook-and-line/ >5,000 snapper line 71 Lewis Fishing not applicable Antigua & Barbuda EEZ none documented none documented N/I, A/G U.S. -

Betanodavirus and VER Disease: a 30-Year Research Review
pathogens Review Betanodavirus and VER Disease: A 30-year Research Review Isabel Bandín * and Sandra Souto Departamento de Microbioloxía e Parasitoloxía-Instituto de Acuicultura, Universidade de Santiago de Compostela, 15782 Santiago de Compostela, Spain; [email protected] * Correspondence: [email protected] Received: 20 December 2019; Accepted: 4 February 2020; Published: 9 February 2020 Abstract: The outbreaks of viral encephalopathy and retinopathy (VER), caused by nervous necrosis virus (NNV), represent one of the main infectious threats for marine aquaculture worldwide. Since the first description of the disease at the end of the 1980s, a considerable amount of research has gone into understanding the mechanisms involved in fish infection, developing reliable diagnostic methods, and control measures, and several comprehensive reviews have been published to date. This review focuses on host–virus interaction and epidemiological aspects, comprising viral distribution and transmission as well as the continuously increasing host range (177 susceptible marine species and epizootic outbreaks reported in 62 of them), with special emphasis on genotypes and the effect of global warming on NNV infection, but also including the latest findings in the NNV life cycle and virulence as well as diagnostic methods and VER disease control. Keywords: nervous necrosis virus (NNV); viral encephalopathy and retinopathy (VER); virus–host interaction; epizootiology; diagnostics; control 1. Introduction Nervous necrosis virus (NNV) is the causative agent of viral encephalopathy and retinopathy (VER), otherwise known as viral nervous necrosis (VNN). The disease was first described at the end of the 1980s in Australia and in the Caribbean [1–3], and has since caused a great deal of mortalities and serious economic losses in a variety of reared marine fish species, but also in freshwater species worldwide. -

Bilateral Asymmetry and Bilateral Variation in Fishes *
BILATERAL ASYMMETRY AND BILATERAL VARIATION IN FISHES * bARL L. HUBBS AND LAURA C. HUBBS CONTENTS PAGE Introduction ................................................................................................................... 230 Statistical methods ....................................................................................................... 231 Dextrality and sinistrality in flatfishes .................................................................. 234 Reversal of sides in flounders .............................................................................. 236 Decreased viability of reversed flounders ......................................................... 240 Incomplete mirror imaging in reversed flounders .......................................... 243 A completely reversed flatfish .............................................................................. 245 Interpretation of reversal in flatfishes ............................................................... 246 Teratological return toward symmetry ............................................................. 249 Secondary asymmetries in flatfishes .................................................................... 250 Bilateral variation in number of rays in paired fins on the two sides of flatfishes ................................................................................................................. 254 Asymmetries and bilateral variations in essentially symmetrical fishes ....... 263 Bilateral variation in number of rays in the left -

Sperm Cryopreservation
animals Article Effects of Cryoprotective Medium Composition, Dilution Ratio, and Freezing Rates on Spotted Halibut (Verasper variegatus) Sperm Cryopreservation Irfan Zidni 1 , Yun Ho Lee 1, Jung Yeol Park 1, Hyo Bin Lee 1, Jun Wook Hur 2 and Han Kyu Lim 1,* 1 Department of Marine and Fisheries Resources, Mokpo National University, Mokpo 58554, Korea; [email protected] (I.Z.); [email protected] (Y.H.L.); [email protected] (J.Y.P.); [email protected] (H.B.L.) 2 Faculty of Marine Applied Biosciences, Kunsan National University, 558 Daehak-ro, Gunsan, Jeonbuk 54150, Korea; [email protected] * Correspondence: [email protected]; Tel.: +82-61-450-2395; Fax: +82-61-452-8875 Received: 12 October 2020; Accepted: 16 November 2020; Published: 19 November 2020 Simple Summary: The spotted halibut, Verasper variegatus, is a popular fish species occurring naturally in the East China Sea and coastal areas of Korea and Japan. However, when reared in captivity, male and female spotted halibut do not usually mature synchronously. Maintaining production of this commercial fish in hatcheries through sperm cryopreservation is important. This study investigated the effect of several factors for successful cryopreservation of fish sperm including cryoprotective agents (CPAs), diluents, dilution ratios (Milt: CPA + diluents), and freezing rates. The observed factors significantly affected movable sperm ratio (MSR), sperm activity index (SAI), survival rate, and DNA damage after cryopreservation. In the present study, the mixture of 15% dimethyl sulfoxide (DMSO) + 300 mM sucrose with a dilution ratio lower than 1:2 and a freezing rate slower than 5 C/min provided the best treatment and reduced DNA damage.