Locus Number Estimation of MHC Class II B in Stone Flounder and Japanese Flounder
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Winter Flounder Pseudopleuronectes Americanus Stock Enhancement in New Hampshire: Developing Optimal Release Strategies
University of New Hampshire University of New Hampshire Scholars' Repository Doctoral Dissertations Student Scholarship Spring 2002 Winter flounder Pseudopleuronectes americanus stock enhancement in New Hampshire: Developing optimal release strategies Elizabeth Alden Fairchild University of New Hampshire, Durham Follow this and additional works at: https://scholars.unh.edu/dissertation Recommended Citation Fairchild, Elizabeth Alden, "Winter flounder Pseudopleuronectes americanus stock enhancement in New Hampshire: Developing optimal release strategies" (2002). Doctoral Dissertations. 62. https://scholars.unh.edu/dissertation/62 This Dissertation is brought to you for free and open access by the Student Scholarship at University of New Hampshire Scholars' Repository. It has been accepted for inclusion in Doctoral Dissertations by an authorized administrator of University of New Hampshire Scholars' Repository. For more information, please contact [email protected]. INFORMATION TO USERS This manuscript has been reproduced from the microfilm master. UMI films the text directly from the original or copy submitted. Thus, some thesis and dissertation copies are in typewriter face, while others may be from any type of computer printer. The quality of this reproduction is dependent upon the quality of the copy submitted. Broken or indistinct print, colored or poor quality illustrations and photographs, print bleedthrough, substandard margins, and improper alignment can adversely affect reproduction. In the unlikely event that the author did not send UMI a complete manuscript and there are missing pages, these will be noted. Also, if unauthorized copyright material had to be removed, a note will indicate the deletion. Oversize materials (e.g., maps, drawings, charts) are reproduced by sectioning the original, beginning at the upper left-hand comer and continuing from left to right in equal sections with small overlaps. -

Disease List for Aquaculture Health Certificate
Quarantine Standard for Designated Species of Imported/Exported Aquatic Animals [Attached Table] 4. Listed Diseases & Quarantine Standard for Designated Species Listed disease designated species standard Common name Disease Pathogen 1. Epizootic haematopoietic Epizootic Perca fluviatilis Redfin perch necrosis(EHN) haematopoietic Oncorhynchus mykiss Rainbow trout necrosis virus(EHNV) Macquaria australasica Macquarie perch Bidyanus bidyanus Silver perch Gambusia affinis Mosquito fish Galaxias olidus Mountain galaxias Negative Maccullochella peelii Murray cod Salmo salar Atlantic salmon Ameirus melas Black bullhead Esox lucius Pike 2. Spring viraemia of Spring viraemia of Cyprinus carpio Common carp carp, (SVC) carp virus(SVCV) Grass carp, Ctenopharyngodon idella white amur Hypophthalmichthys molitrix Silver carp Hypophthalmichthys nobilis Bighead carp Carassius carassius Crucian carp Carassius auratus Goldfish Tinca tinca Tench Sheatfish, Silurus glanis European catfish, wels Negative Leuciscus idus Orfe Rutilus rutilus Roach Danio rerio Zebrafish Esox lucius Northern pike Poecilia reticulata Guppy Lepomis gibbosus Pumpkinseed Oncorhynchus mykiss Rainbow trout Abramis brama Freshwater bream Notemigonus cysoleucas Golden shiner 3.Viral haemorrhagic Viral haemorrhagic Oncorhynchus spp. Pacific salmon septicaemia(VHS) septicaemia Oncorhynchus mykiss Rainbow trout virus(VHSV) Gadus macrocephalus Pacific cod Aulorhynchus flavidus Tubesnout Cymatogaster aggregata Shiner perch Ammodytes hexapterus Pacific sandlance Merluccius productus Pacific -
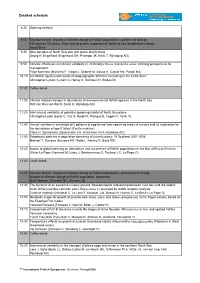
Schedule Onlinepdf
Detailed schedule 8:30 Opening session 9:00 Keynote lecture: Impacts of climate change on flatfish populations - patterns of change 100 days to 100 years: Short and long-term responses of flatfish to sea temperature change David Sims 9:30 Nine decades of North Sea sole and plaice distributions Georg H. Engelhard (Engelhard GH, Pinnegar JK, Kell LT, Rijnsdorp AD) 9:50 Climatic effects on recruitment variability in Platichthys flesus and Solea solea: defining perspectives for management. Filipe Martinho (Martinho F, Viegas I, Dolbeth M, Sousa H, Cabral HN, Pardal MA) 10:10 Are flatfish species with southern biogeographic affinities increasing in the Celtic Sea? Christopher Lynam (Lynam C, Harlay X, Gerritsen H, Stokes D) 10:30 Coffee break 11:00 Climate related changes in abundance of non-commercial flatfish species in the North Sea Ralf van Hal (van Hal R, Smits K, Rijnsdorp AD) 11:20 Inter-annual variability of potential spawning habitat of North Sea plaice Christophe Loots (Loots C, Vaz S, Koubii P, Planque B, Coppin F, Verin Y) 11:40 Annual variation in simulated drift patterns of egg/larvae from spawning areas to nursery and its implication for the abundance of age-0 turbot (Psetta maxima) Claus R. Sparrevohn (Sparrevohn CR, Hinrichsen H-H, Rijnsdorp AD) 12:00 Broadscale patterns in population dynamics of juvenile plaice: W Scotland 2001-2008 Michael T. Burrows (Burrows MT, Robb L, Harvey R, Batty RS) 12:20 Impact of global warming on abundance and occurrence of flatfish populations in the Bay of Biscay (France) Olivier Le Pape (Hermant -

(Paralichthys Lethostigma) in the Galveston Bay Estuary, TX
DISTRIBUTION, CONDITION, AND GROWTH OF NEWLY SETTLED SOUTHERN FLOUNDER (Paralichthys lethostigma) IN THE GALVESTON BAY ESTUARY, TX A Thesis by LINDSAY ANN GLASS Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE May 2006 Major Subject: Wildlife and Fisheries Sciences DISTRIBUTION, CONDITION, AND GROWTH OF NEWLY SETTLED SOUTHERN FLOUNDER (Paralichthys lethostigma) IN THE GALVESTON BAY ESTUARY, TX A Thesis by LINDSAY ANN GLASS Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE Approved by: Chair of Committee, Jay R. Rooker Committee Members, William H. Neill Antonietta Quigg Head of Department, Delbert M.Gatlin III May 2006 Major Subject: Wildlife and Fisheries Sciences iii ABSTRACT Distribution, Condition, and Growth of Newly Settled Southern Flounder (Paralichthys lethostigma) in the Galveston Bay Estuary, TX. (May 2006) Lindsay Ann Glass, B.S., Texas A&M University-Galveston Chair of Advisory Committee: Dr. Jay R. Rooker Several flatfish species including southern flounder (Paralichthys lethostigma) recruit to estuaries during early life. Therefore, the evaluation of estuarine sites and habitats that serve as nurseries is critical to conservation and management efforts. I used biochemical condition and growth measurements in conjunction with catch-density data to evaluate settlement sites used by southern flounder in the Galveston Bay Estuary (GBE). In 2005, beam-trawl collections were made in three major sections of the GBE (East Bay, West Bay, Galveston Bay), and three sites were sampled in each bay. -

Distribution and Abundance of Pleuronectiformes Larvae Off Southeastern Brazil
BRAZILIAN JOURNAL OF OCEANOGRAPHY, 62(1):23-34, 2014 DISTRIBUTION AND ABUNDANCE OF PLEURONECTIFORMES LARVAE OFF SOUTHEASTERN BRAZIL Camilla Nunes Garbini*, Maria de Lourdes Zani-Teixeira , Márcio Hidekazu Ohkawara and Mario Katsuragawa Instituto Oceanográfico da Universidade de São Paulo (Praça do Oceanográfico, 191, 05508-120 São Paulo, SP, Brasil) *Corresponding author: [email protected] http://dx.doi.org/10.1590/S1679-87592014051706201 ABSTRACT The objective of this study was the description of the composition, abundance and density in horizontal and vertical distribution of Pleuronectiformes larvae on the southeastern Brazilian continental shelf. The samples were collected with bongo nets and a Multi Plankton Sampler (MPS), both in summer and winter 2002. A total of 352 flatfishes larvae were collected in summer and 343 in winter, representing three families and a total of 13 taxa: Paralichthyidae ( Citharichthys cornutus, C. spilopterus, Citharichthys sp ., Cyclopsetta chittendeni, Syacium spp ., Etropus spp . and Paralichthys spp .), Bothidae ( Bothus ocellatus and Monolene antillarum ) and Cynoglossidae ( Symphurus trewavasae, S. jenynsi, S. plagusia and S. ginsburgi ). The most abundant taxa were Etropus spp ., Syacium spp . and Bothus ocellatus . Etropus spp . occurred mainly as far out as the 200 m isobath and Syacium spp . from 100 m. B. ocellatus was present mainly in the oceanic zone between Ubatuba and Rio de Janeiro as from the 200 m isobath. The greatest average densities of these species occurred in the strata from 0 to 20 m depth in summer and between 20 and 40 m in winter. RESUMO O objetivo deste estudo foi descrever a composição, abundância, densidade, distribuição horizontal e vertical das larvas de Pleuronectiformes ao longo da plataforma continental Sudeste brasileira. -

Sperm Cryopreservation
animals Article Effects of Cryoprotective Medium Composition, Dilution Ratio, and Freezing Rates on Spotted Halibut (Verasper variegatus) Sperm Cryopreservation Irfan Zidni 1 , Yun Ho Lee 1, Jung Yeol Park 1, Hyo Bin Lee 1, Jun Wook Hur 2 and Han Kyu Lim 1,* 1 Department of Marine and Fisheries Resources, Mokpo National University, Mokpo 58554, Korea; [email protected] (I.Z.); [email protected] (Y.H.L.); [email protected] (J.Y.P.); [email protected] (H.B.L.) 2 Faculty of Marine Applied Biosciences, Kunsan National University, 558 Daehak-ro, Gunsan, Jeonbuk 54150, Korea; [email protected] * Correspondence: [email protected]; Tel.: +82-61-450-2395; Fax: +82-61-452-8875 Received: 12 October 2020; Accepted: 16 November 2020; Published: 19 November 2020 Simple Summary: The spotted halibut, Verasper variegatus, is a popular fish species occurring naturally in the East China Sea and coastal areas of Korea and Japan. However, when reared in captivity, male and female spotted halibut do not usually mature synchronously. Maintaining production of this commercial fish in hatcheries through sperm cryopreservation is important. This study investigated the effect of several factors for successful cryopreservation of fish sperm including cryoprotective agents (CPAs), diluents, dilution ratios (Milt: CPA + diluents), and freezing rates. The observed factors significantly affected movable sperm ratio (MSR), sperm activity index (SAI), survival rate, and DNA damage after cryopreservation. In the present study, the mixture of 15% dimethyl sulfoxide (DMSO) + 300 mM sucrose with a dilution ratio lower than 1:2 and a freezing rate slower than 5 C/min provided the best treatment and reduced DNA damage. -
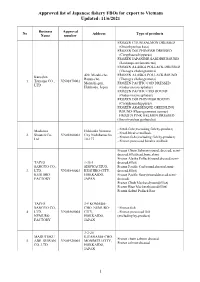
Approved List of Japanese Fishery Fbos for Export to Vietnam Updated: 11/6/2021
Approved list of Japanese fishery FBOs for export to Vietnam Updated: 11/6/2021 Business Approval No Address Type of products Name number FROZEN CHUM SALMON DRESSED (Oncorhynchus keta) FROZEN DOLPHINFISH DRESSED (Coryphaena hippurus) FROZEN JAPANESE SARDINE ROUND (Sardinops melanostictus) FROZEN ALASKA POLLACK DRESSED (Theragra chalcogramma) 420, Misaki-cho, FROZEN ALASKA POLLACK ROUND Kaneshin Rausu-cho, (Theragra chalcogramma) 1. Tsuyama CO., VN01870001 Menashi-gun, FROZEN PACIFIC COD DRESSED LTD Hokkaido, Japan (Gadus macrocephalus) FROZEN PACIFIC COD ROUND (Gadus macrocephalus) FROZEN DOLPHIN FISH ROUND (Coryphaena hippurus) FROZEN ARABESQUE GREENLING ROUND (Pleurogrammus azonus) FROZEN PINK SALMON DRESSED (Oncorhynchus gorbuscha) - Fresh fish (excluding fish by-product) Maekawa Hokkaido Nemuro - Fresh bivalve mollusk. 2. Shouten Co., VN01860002 City Nishihamacho - Frozen fish (excluding fish by-product) Ltd 10-177 - Frozen processed bivalve mollusk Frozen Chum Salmon (round, dressed, semi- dressed,fillet,head,bone,skin) Frozen Alaska Pollack(round,dressed,semi- TAIYO 1-35-1 dressed,fillet) SANGYO CO., SHOWACHUO, Frozen Pacific Cod(round,dressed,semi- 3. LTD. VN01840003 KUSHIRO-CITY, dressed,fillet) KUSHIRO HOKKAIDO, Frozen Pacific Saury(round,dressed,semi- FACTORY JAPAN dressed) Frozen Chub Mackerel(round,fillet) Frozen Blue Mackerel(round,fillet) Frozen Salted Pollack Roe TAIYO 3-9 KOMABA- SANGYO CO., CHO, NEMURO- - Frozen fish 4. LTD. VN01860004 CITY, - Frozen processed fish NEMURO HOKKAIDO, (excluding by-product) FACTORY JAPAN -

Rancangan Keputusan Menteri Kelautan Dan
RANCANGAN KEPUTUSAN MENTERI KELAUTAN DAN PERIKANAN REPUBLIK INDONESIA NOMOR /KEPMEN-KP/2018 TENTANG PENETAPAN JENIS-JENIS HAMA DAN PENYAKIT IKAN KARANTINA, GOLONGAN, DAN MEDIA PEMBAWA DENGAN RAHMAT TUHAN YANG MAHA ESA MENTERI KELAUTAN DAN PERIKANAN REPUBLIK INDONESIA, Menimbang : a. bahwa dengan semakin berkembangnya jenis-jenis hama dan penyakit ikan karantina di luar negeri, serta dalam rangka pelaksanaan pencegahan dan pengendalian penyebaran hama dan penyakit ikan karantina, perlu meninjau kembali Keputusan Menteri Kelautan dan Perikanan Nomor 80/KEPMEN-KP/2015 tentang Penetapan Jenis-Jenis Hama dan Penyakit Ikan Karantina, Golongan, Media Pembawa dan Sebarannya; b. bahwa untuk itu perlu menetapkan Keputusan Menteri Kelautan dan Perikanan tentang Penetapan Jenis-Jenis Hama dan Penyakit Ikan Karantina, Golongan, dan Media Pembawa; Mengingat : 1. Undang-Undang Nomor 16 Tahun 1992 tentang Karantina Hewan, Ikan, dan Tumbuhan (Lembaran Negara Republik Indonesia Tahun 1992 Nomor 56, Tambahan Lembaran Negara Nomor 3482); 2. Peraturan Pemerintah Nomor 15 Tahun 2002 tentang Karantina Ikan (Lembaran Negara Republik Indonesia Tahun 2002 Nomor 36, Tambahan Lembaran Negara Nomor 4197); 3. Peraturan Presiden Nomor 7 Tahun 2015 tentang Organisasi Kementerian Negara (Lembaran Negara Republik Indonesia Tahun 2015 Nomor 8); 4. Peraturan Presiden Nomor Nomor 63 Tahun 2015 tentang Kementerian Kelautan dan Perikanan (Lembaran Negara Republik Indonesia Tahun 2015 Nomor 111), sebagaimana telah diubah dengan Peraturan Presiden Nomor 2 Tahun 2017 tentang -

Quarantine Standard for Designated Species of Imported/Exported Aquatic Animals
Quarantine Standard for Designated Species of Imported/Exported Aquatic Animals [Attached Table] 4. Listed Diseases & Quarantine Standard for Designated Species Listed disease designated species standard Common name Disease Pathogen 1. Epizootic haematopoietic Epizootic Perca fluviatilis Redfin perch necrosis(EHN) haematopoietic Oncorhynchus mykiss Rainbow trout necrosis virus(EHNV) Macquaria australasica Macquarie perch Bidyanus bidyanus Silver perch Gambusia affinis Mosquito fish Galaxias olidus Mountain galaxias Negative Maccullochella peelii Murray cod Salmo salar Atlantic salmon Ameirus melas Black bullhead Esox lucius Pike 2. Spring viraemia of Spring viraemia of Cyprinus carpio Common carp carp, (SVC) carp virus(SVCV) Grass carp, Ctenopharyngodon idella white amur Hypophthalmichthys molitrix Silver carp Hypophthalmichthys nobilis Bighead carp Carassius carassius Crucian carp Carassius auratus Goldfish Tinca tinca Tench Sheatfish, Silurus glanis European catfish, wels Negative Leuciscus idus Orfe Rutilus rutilus Roach Danio rerio Zebrafish Esox lucius Northern pike Poecilia reticulata Guppy Lepomis gibbosus Pumpkinseed Oncorhynchus mykiss Rainbow trout Abramis brama Freshwater bream Notemigonus cysoleucas Golden shiner 3.Viral haemorrhagic Viral haemorrhagic Oncorhynchus spp. Pacific salmon septicaemia(VHS) septicaemia Oncorhynchus mykiss Rainbow trout virus(VHSV) Gadus macrocephalus Pacific cod Aulorhynchus flavidus Tubesnout Cymatogaster aggregata Shiner perch Ammodytes hexapterus Pacific sandlance Merluccius productus Pacific -

The United States and Japanese Counterpart Panels on Aquaculture
PREFACE TheUnited States and Japanese counterpart panels onaquaculture wereformed in 1969 under the UnitedStates-Japan Cooperative Program inNatural Resources UJNR! The panels currently include specialistsdrawnfrom the government andacadeinic departinents mostconcerned withaquaculture- Chargedwith exploring anddeveloping bilateral cooperation, thepanels have focused their efforts on exchanginginformation related to aquaculture thatcould be of benefittoboth countries. TheUJNR was begun during theThird Cabinet Level Meeting ofthe Joint United States-Japan CommitteeonTrade and Economic Affairsin January 1964,ln addition toaquaculture, currentsubjects in theprogram include toxicmicroorganisms, airpollution, energy, forage crops, national parkmanagement, rnycoplasmosis,windand seismic effects, protein resources, forestry, andseveral joint panels and corninitteesin marine resolve research, development, andutilization. Accomplishmentsiticlude:increased communication andcooperation amongtechnical specialists; exchangesofinformation, data,and research findings; annual meetings ofthe panel, a policy-coordinating body;administrative staffmeetings; exchanges ofequipment, materials, andsamples; several major technicalconferences; andbeneficial effects of internationalrelations. The26th U.S.-Japan Aquaculture PanelSyinposium washeld in Durham, New Hampshire, from16- 18September ] 997. Following thesyrnposiuin, fieldtrips during a seven-day period included theareas of Portsmouth,NewHainpshire; andBar Harbor, Eastport, Camden, and Boothbay Harbor, Maine. -

Euryhalinity in an Evolutionary Context Eric T
University of Connecticut OpenCommons@UConn EEB Articles Department of Ecology and Evolutionary Biology 2013 Euryhalinity in an Evolutionary Context Eric T. Schultz University of Connecticut - Storrs, [email protected] Stephen D. McCormick USGS Conte Anadromous Fish Research Center, [email protected] Follow this and additional works at: https://opencommons.uconn.edu/eeb_articles Part of the Comparative and Evolutionary Physiology Commons, Evolution Commons, and the Terrestrial and Aquatic Ecology Commons Recommended Citation Schultz, Eric T. and McCormick, Stephen D., "Euryhalinity in an Evolutionary Context" (2013). EEB Articles. 29. https://opencommons.uconn.edu/eeb_articles/29 RUNNING TITLE: Evolution and Euryhalinity Euryhalinity in an Evolutionary Context Eric T. Schultz Department of Ecology and Evolutionary Biology, University of Connecticut Stephen D. McCormick USGS, Conte Anadromous Fish Research Center, Turners Falls, MA Department of Biology, University of Massachusetts, Amherst Corresponding author (ETS) contact information: Department of Ecology and Evolutionary Biology University of Connecticut Storrs CT 06269-3043 USA [email protected] phone: 860 486-4692 Keywords: Cladogenesis, diversification, key innovation, landlocking, phylogeny, salinity tolerance Schultz and McCormick Evolution and Euryhalinity 1. Introduction 2. Diversity of halotolerance 2.1. Empirical issues in halotolerance analysis 2.2. Interspecific variability in halotolerance 3. Evolutionary transitions in euryhalinity 3.1. Euryhalinity and halohabitat transitions in early fishes 3.2. Euryhalinity among extant fishes 3.3. Evolutionary diversification upon transitions in halohabitat 3.4. Adaptation upon transitions in halohabitat 4. Convergence and euryhalinity 5. Conclusion and perspectives 2 Schultz and McCormick Evolution and Euryhalinity This chapter focuses on the evolutionary importance and taxonomic distribution of euryhalinity. Euryhalinity refers to broad halotolerance and broad halohabitat distribution. -

Tracing the Genetic Impact of Farmed Turbot Scophthalmus Maximus on Wild Populations
Vol. 10: 447–463, 2018 AQUACULTURE ENVIRONMENT INTERACTIONS Published November 1 https://doi.org/10.3354/aei00282 Aquacult Environ Interact OPENPEN ACCESSCCESS Tracing the genetic impact of farmed turbot Scophthalmus maximus on wild populations F. D. Prado1,2, M. Vera1, M. Hermida1, A. Blanco1, C. Bouza1, G. E. Maes3,4, F. A. M. Volckaert3, AquaTrace Consortium5, P. Martínez1,* 1Department of Zoology, Genetics and Physical Anthropology, Faculty of Veterinary, Universidade de Santiago de Compostela, 27002 Lugo, Spain 2CAPES Foundation, Ministry of Education of Brazil, Brasília, Brazil 3Laboratory of Biodiversity and Evolutionary Genomics, KU Leuven, 3000 Leuven, Belgium 4Laboratory for Cytogenetics and Genome Research, Center for Human Genetics, Genomics Core, KU Leuven, 3000 Leuven, Belgium 5European FP7 KBBE project AquaTrace (311920): the development of tools for tracing and evaluating the genetic impact of fish from aquaculture ABSTRACT: The impact of escapees from aquaculture is of general concern for the sustainability of natural resources. Turbot Scophthalmus maximus is a marine flatfish of great commercial value whose land-based aquaculture started approx. 40 yr ago; hence, a low impact of escapees is expected on wild populations. However, enhancement of wild stocks using farmed turbot has been carried out along the Northeast Atlantic coasts in the last decades. Recently, a broad panel of single nucleotide polymorphism (SNP) markers (755 SNPs; 1 SNP Mb−1) has been used to eval- uate the genetic structure of turbot throughout its distribution range, constituting the baseline to evaluate the impact of farmed fish in the wild. Two distinct origins were identified for farmed tur- bot (F_ORI1 and F_ORI2; FST = 0.049), which differentiated from wild populations after 5 generations of selection (average FST = 0.059), and consistent evidence of adaptation to domestication was de - tected.