Tracing the Genetic Impact of Farmed Turbot Scophthalmus Maximus on Wild Populations
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Disease List for Aquaculture Health Certificate
Quarantine Standard for Designated Species of Imported/Exported Aquatic Animals [Attached Table] 4. Listed Diseases & Quarantine Standard for Designated Species Listed disease designated species standard Common name Disease Pathogen 1. Epizootic haematopoietic Epizootic Perca fluviatilis Redfin perch necrosis(EHN) haematopoietic Oncorhynchus mykiss Rainbow trout necrosis virus(EHNV) Macquaria australasica Macquarie perch Bidyanus bidyanus Silver perch Gambusia affinis Mosquito fish Galaxias olidus Mountain galaxias Negative Maccullochella peelii Murray cod Salmo salar Atlantic salmon Ameirus melas Black bullhead Esox lucius Pike 2. Spring viraemia of Spring viraemia of Cyprinus carpio Common carp carp, (SVC) carp virus(SVCV) Grass carp, Ctenopharyngodon idella white amur Hypophthalmichthys molitrix Silver carp Hypophthalmichthys nobilis Bighead carp Carassius carassius Crucian carp Carassius auratus Goldfish Tinca tinca Tench Sheatfish, Silurus glanis European catfish, wels Negative Leuciscus idus Orfe Rutilus rutilus Roach Danio rerio Zebrafish Esox lucius Northern pike Poecilia reticulata Guppy Lepomis gibbosus Pumpkinseed Oncorhynchus mykiss Rainbow trout Abramis brama Freshwater bream Notemigonus cysoleucas Golden shiner 3.Viral haemorrhagic Viral haemorrhagic Oncorhynchus spp. Pacific salmon septicaemia(VHS) septicaemia Oncorhynchus mykiss Rainbow trout virus(VHSV) Gadus macrocephalus Pacific cod Aulorhynchus flavidus Tubesnout Cymatogaster aggregata Shiner perch Ammodytes hexapterus Pacific sandlance Merluccius productus Pacific -
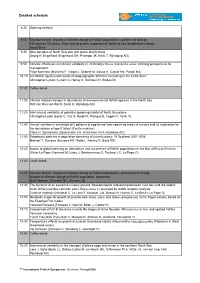
Schedule Onlinepdf
Detailed schedule 8:30 Opening session 9:00 Keynote lecture: Impacts of climate change on flatfish populations - patterns of change 100 days to 100 years: Short and long-term responses of flatfish to sea temperature change David Sims 9:30 Nine decades of North Sea sole and plaice distributions Georg H. Engelhard (Engelhard GH, Pinnegar JK, Kell LT, Rijnsdorp AD) 9:50 Climatic effects on recruitment variability in Platichthys flesus and Solea solea: defining perspectives for management. Filipe Martinho (Martinho F, Viegas I, Dolbeth M, Sousa H, Cabral HN, Pardal MA) 10:10 Are flatfish species with southern biogeographic affinities increasing in the Celtic Sea? Christopher Lynam (Lynam C, Harlay X, Gerritsen H, Stokes D) 10:30 Coffee break 11:00 Climate related changes in abundance of non-commercial flatfish species in the North Sea Ralf van Hal (van Hal R, Smits K, Rijnsdorp AD) 11:20 Inter-annual variability of potential spawning habitat of North Sea plaice Christophe Loots (Loots C, Vaz S, Koubii P, Planque B, Coppin F, Verin Y) 11:40 Annual variation in simulated drift patterns of egg/larvae from spawning areas to nursery and its implication for the abundance of age-0 turbot (Psetta maxima) Claus R. Sparrevohn (Sparrevohn CR, Hinrichsen H-H, Rijnsdorp AD) 12:00 Broadscale patterns in population dynamics of juvenile plaice: W Scotland 2001-2008 Michael T. Burrows (Burrows MT, Robb L, Harvey R, Batty RS) 12:20 Impact of global warming on abundance and occurrence of flatfish populations in the Bay of Biscay (France) Olivier Le Pape (Hermant -

04-Bailly 669.Indd
Scophthalmus Rafinesque, 1810: The valid generic name for the turbot, S. maximus (Linnaeus, 1758) [Pleuronectiformes: Scophthalmidae] by Nicolas BAILLY* (1) & Bruno CHANET (2) ABSTRACT. - In the past 50 years, the turbot is referred to either as Scophthalmus maximus (Linnaeus, 1758) or Psetta maxima (Linnaeus, 1758) in the literature. Norman (1931) had argued that the valid name for the turbot was Scophthalmus maximus. However, his recommendation was never universally accepted, and today the confusing situation exists where two generic names are still being used for this species. We address this issue by analysing findings from recently published works on the anatomy, molecular and morphological phylogenetic systematics, and ecology of scophthalmid fishes. The preponderance of evidence supports the strong recommendation to use Scophthalmus as the valid generic name for the tur- bot. Acceptance of this generic name conveys the best information available concerning the systematic relationships of this species, and also serves to simplify the nomenclature of scophthalmid flatfishes in publications on systematics, fisheries and aquaculture, fishery statistics, ichthyofaunal and field guides for the general public, and in various legal and conserva- tion-related documents. This paper reinforces the conclusions of Chanet (2003) with more arguments. RÉSUMÉ. - Scophthalmus Rafinesque, 1810: le nom de genre valide du turbot,S. maximus (Linnaeus, 1758) (Pleuronecti- formes: Scophthalmidae). Depuis 50 ans, le turbot est dénommé dans la littérature soit Scophthalmus maximus (Linnaeus, 1758), soit Psetta maxima (Linnaeus, 1758). Norman (1931) avait montré que le nom valide pour le turbot était Scophthalmus maximus. Cependant, sa recommandation ne fut jamais universellement appliquée, et aujourd’hui la situation reste confuse avec deux noms génériques en usage pour cette espèce. -

Food Choice of Different Size Classes of Flounder (Platichthys Flesus ) In
Food choice of different size classes of flounder ( Platichthys flesus ) in the Baltic Sea Jennie Ljungberg Degree project in biology, Master of science (2 years), 2014 Examensarbete i biologi 30 hp till masterexamen, 2014 Biology Education Centre Supervisor: Bertil Widbom Table of Contents ABSTRACT ............................................................................................................................................ 3 INTRODUCTION ................................................................................................................................... 4 Flounders in the Baltic Sea .................................................................................................................. 5 The diet of flounders ........................................................................................................................... 6 Blue mussel (Mytilus edulis) ............................................................................................................... 7 Blue mussels in the Baltic Sea............................................................................................................. 8 The nutritive value of blue mussels ..................................................................................................... 9 The condition of flounders in the Baltic Sea ....................................................................................... 9 Aims ................................................................................................................................................. -

Locus Number Estimation of MHC Class II B in Stone Flounder and Japanese Flounder
Int. J. Mol. Sci. 2015, 16, 6000-6017; doi:10.3390/ijms16036000 OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Article Locus Number Estimation of MHC Class II B in Stone Flounder and Japanese Flounder Jiajun Jiang †, Chunmei Li †, Quanqi Zhang and Xubo Wang * Key Laboratory of Marine Genetics and Breeding (Ocean University of China), Ministry of Education, Qingdao 266003, China; E-Mails: [email protected] (J.J.); [email protected] (C.L.); [email protected] (Q.Z.) † These authors contributed equally to this work. * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel./Fax: +86-532-8203-1986. Academic Editor: Ritva Tikkanen Received: 2 December 2014 / Accepted: 25 December 2014 / Published: 13 March 2015 Abstract: Members of major histocompatibility complex (MHC) family are important in immune systems. Great efforts have been made to reveal their complicated gene structures. But many existing studies focus on partial sequences of MHC genes. In this study, by gene cloning and sequencing, we identified cDNA sequences and DNA sequences of the MHC class II B in two flatfishes, stone flounder (Kareius bicoloratus) and homozygous diploid Japanese flounder (Paralichthys olivaceus). Eleven cDNA sequences were acquired from eight stone flounder individuals, and most of the polymorphic sites distributed in exons 2 and 3. Twenty-eight alleles were identified from the DNA fragments in these eight individuals. It could be deduced from their Bayesian inference phylogenetic tree that at least four loci of MHC class II B exist in stone flounder. The detailed whole-length DNA sequences in one individual were analyzed, revealing that the intron length varied among different loci. -

Atlas of North Sea Fishes
ICES COOPERATIVE RESEARCH REPORT RAPPORT DES RECHERCHES COLLECTIVES NO. 194 Atlas of North Sea Fishes Based on bottom-trawl survey data for the years 1985—1987 Ruud J. Knijn1, Trevor W. Boon2, Henk J. L. Heessen1, and John R. G. Hislop3 'Netherlands Institute for Fisheries Research, Haringkade 1, PO Box 6 8 , 1970 AB Umuiden, The Netherlands 2MAFF, Fisheries Laboratory, Lowestoft, Suffolk NR33 OHT, England 3Marine Laboratory, PO Box 101, Victoria Road, Aberdeen AB9 8 DB, Scotland Fish illustrations by Peter Stebbing International Council for the Exploration of the Sea Conseil International pour l’Exploration de la Mer Palægade 2—4, DK-1261 Copenhagen K, Denmark September 1993 Copyright ® 1993 All rights reserved No part of this book may be reproduced in any form by photostat or microfilm or stored in a storage system or retrieval system or by any other means without written permission from the authors and the International Council for the Exploration of the Sea Illustrations ® 1993 Peter Stebbing Published with financial support from the Directorate-General for Fisheries, AIR Programme, of the Commission of the European Communities ICES Cooperative Research Report No. 194 Atlas of North Sea Fishes ISSN 1017-6195 Printed in Denmark Contents 1. Introduction............................................................................................................... 1 2. Recruit surveys.................................................................................. 3 2.1 General purpose of the surveys..................................................................... -

Sperm Cryopreservation
animals Article Effects of Cryoprotective Medium Composition, Dilution Ratio, and Freezing Rates on Spotted Halibut (Verasper variegatus) Sperm Cryopreservation Irfan Zidni 1 , Yun Ho Lee 1, Jung Yeol Park 1, Hyo Bin Lee 1, Jun Wook Hur 2 and Han Kyu Lim 1,* 1 Department of Marine and Fisheries Resources, Mokpo National University, Mokpo 58554, Korea; [email protected] (I.Z.); [email protected] (Y.H.L.); [email protected] (J.Y.P.); [email protected] (H.B.L.) 2 Faculty of Marine Applied Biosciences, Kunsan National University, 558 Daehak-ro, Gunsan, Jeonbuk 54150, Korea; [email protected] * Correspondence: [email protected]; Tel.: +82-61-450-2395; Fax: +82-61-452-8875 Received: 12 October 2020; Accepted: 16 November 2020; Published: 19 November 2020 Simple Summary: The spotted halibut, Verasper variegatus, is a popular fish species occurring naturally in the East China Sea and coastal areas of Korea and Japan. However, when reared in captivity, male and female spotted halibut do not usually mature synchronously. Maintaining production of this commercial fish in hatcheries through sperm cryopreservation is important. This study investigated the effect of several factors for successful cryopreservation of fish sperm including cryoprotective agents (CPAs), diluents, dilution ratios (Milt: CPA + diluents), and freezing rates. The observed factors significantly affected movable sperm ratio (MSR), sperm activity index (SAI), survival rate, and DNA damage after cryopreservation. In the present study, the mixture of 15% dimethyl sulfoxide (DMSO) + 300 mM sucrose with a dilution ratio lower than 1:2 and a freezing rate slower than 5 C/min provided the best treatment and reduced DNA damage. -

The Round Goby Genome Provides Insights Into Mechanisms That May Facilitate Biological Invasions
Adrian-Kalchhauser et al. BMC Biology (2020) 18:11 https://doi.org/10.1186/s12915-019-0731-8 RESEARCH ARTICLE Open Access The round goby genome provides insights into mechanisms that may facilitate biological invasions Irene Adrian-Kalchhauser1,2* , Anders Blomberg3†, Tomas Larsson4†, Zuzana Musilova5†, Claire R. Peart6†, Martin Pippel7†, Monica Hongroe Solbakken8†, Jaanus Suurväli9†, Jean-Claude Walser10†, Joanna Yvonne Wilson11†, Magnus Alm Rosenblad3,12†, Demian Burguera5†, Silvia Gutnik13†, Nico Michiels14†, Mats Töpel2†, Kirill Pankov11†, Siegfried Schloissnig15† and Sylke Winkler7† Abstract Background: Theinvasivebenthicroundgoby(Neogobius melanostomus) is the most successful temperate invasive fish and has spread in aquatic ecosystems on both sides of the Atlantic. Invasive species constitute powerful in situ experimental systems to study fast adaptation and directional selection on short ecological timescales and present promising case studies to understand factors involved the impressive ability of some species to colonize novel environments. We seize the unique opportunity presented by the round goby invasion to study genomic substrates potentially involved in colonization success. Results: We report a highly contiguous long-read-based genome and analyze gene families that we hypothesize to relate to the ability of these fish to deal with novel environments. The analyses provide novel insights from the large evolutionary scale to the small species-specific scale. We describe expansions in specific cytochromeP450enzymes,aremarkablydiverse innate immune system, an ancient duplication in red light vision accompanied by red skin fluorescence, evolutionary patterns of epigenetic regulators, and the presence of osmoregulatory genes that may have contributed to the round goby’s capacity to invade cold and salty waters. A recurring theme across all analyzed gene families is gene expansions. -
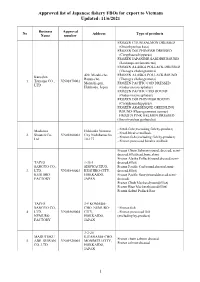
Approved List of Japanese Fishery Fbos for Export to Vietnam Updated: 11/6/2021
Approved list of Japanese fishery FBOs for export to Vietnam Updated: 11/6/2021 Business Approval No Address Type of products Name number FROZEN CHUM SALMON DRESSED (Oncorhynchus keta) FROZEN DOLPHINFISH DRESSED (Coryphaena hippurus) FROZEN JAPANESE SARDINE ROUND (Sardinops melanostictus) FROZEN ALASKA POLLACK DRESSED (Theragra chalcogramma) 420, Misaki-cho, FROZEN ALASKA POLLACK ROUND Kaneshin Rausu-cho, (Theragra chalcogramma) 1. Tsuyama CO., VN01870001 Menashi-gun, FROZEN PACIFIC COD DRESSED LTD Hokkaido, Japan (Gadus macrocephalus) FROZEN PACIFIC COD ROUND (Gadus macrocephalus) FROZEN DOLPHIN FISH ROUND (Coryphaena hippurus) FROZEN ARABESQUE GREENLING ROUND (Pleurogrammus azonus) FROZEN PINK SALMON DRESSED (Oncorhynchus gorbuscha) - Fresh fish (excluding fish by-product) Maekawa Hokkaido Nemuro - Fresh bivalve mollusk. 2. Shouten Co., VN01860002 City Nishihamacho - Frozen fish (excluding fish by-product) Ltd 10-177 - Frozen processed bivalve mollusk Frozen Chum Salmon (round, dressed, semi- dressed,fillet,head,bone,skin) Frozen Alaska Pollack(round,dressed,semi- TAIYO 1-35-1 dressed,fillet) SANGYO CO., SHOWACHUO, Frozen Pacific Cod(round,dressed,semi- 3. LTD. VN01840003 KUSHIRO-CITY, dressed,fillet) KUSHIRO HOKKAIDO, Frozen Pacific Saury(round,dressed,semi- FACTORY JAPAN dressed) Frozen Chub Mackerel(round,fillet) Frozen Blue Mackerel(round,fillet) Frozen Salted Pollack Roe TAIYO 3-9 KOMABA- SANGYO CO., CHO, NEMURO- - Frozen fish 4. LTD. VN01860004 CITY, - Frozen processed fish NEMURO HOKKAIDO, (excluding by-product) FACTORY JAPAN -

Exotic Species in the Aegean, Marmara, Black, Azov and Caspian Seas
EXOTIC SPECIES IN THE AEGEAN, MARMARA, BLACK, AZOV AND CASPIAN SEAS Edited by Yuvenaly ZAITSEV and Bayram ÖZTÜRK EXOTIC SPECIES IN THE AEGEAN, MARMARA, BLACK, AZOV AND CASPIAN SEAS All rights are reserved. No part of this publication may be reproduced, stored in a retrieval system, or transmitted in any form or by any means without the prior permission from the Turkish Marine Research Foundation (TÜDAV) Copyright :Türk Deniz Araştırmaları Vakfı (Turkish Marine Research Foundation) ISBN :975-97132-2-5 This publication should be cited as follows: Zaitsev Yu. and Öztürk B.(Eds) Exotic Species in the Aegean, Marmara, Black, Azov and Caspian Seas. Published by Turkish Marine Research Foundation, Istanbul, TURKEY, 2001, 267 pp. Türk Deniz Araştırmaları Vakfı (TÜDAV) P.K 10 Beykoz-İSTANBUL-TURKEY Tel:0216 424 07 72 Fax:0216 424 07 71 E-mail :[email protected] http://www.tudav.org Printed by Ofis Grafik Matbaa A.Ş. / İstanbul -Tel: 0212 266 54 56 Contributors Prof. Abdul Guseinali Kasymov, Caspian Biological Station, Institute of Zoology, Azerbaijan Academy of Sciences. Baku, Azerbaijan Dr. Ahmet Kıdeys, Middle East Technical University, Erdemli.İçel, Turkey Dr. Ahmet . N. Tarkan, University of Istanbul, Faculty of Fisheries. Istanbul, Turkey. Prof. Bayram Ozturk, University of Istanbul, Faculty of Fisheries and Turkish Marine Research Foundation, Istanbul, Turkey. Dr. Boris Alexandrov, Odessa Branch, Institute of Biology of Southern Seas, National Academy of Ukraine. Odessa, Ukraine. Dr. Firdauz Shakirova, National Institute of Deserts, Flora and Fauna, Ministry of Nature Use and Environmental Protection of Turkmenistan. Ashgabat, Turkmenistan. Dr. Galina Minicheva, Odessa Branch, Institute of Biology of Southern Seas, National Academy of Ukraine. -

Threatened Brill Species in Marine Waters of Turkey: Scopthalmus Rhombus (Linnaeus, 1758) (Scopthalmidae)
Natural and Engineering Sciences 1 Volume 1, No. 1, 1-6, 2016 Threatened brill species in marine waters of Turkey: Scopthalmus rhombus (Linnaeus, 1758) (Scopthalmidae) Cemal Turan1*, Deniz Yağlioğlu2, Deniz Ergüden1, Mevlüt Gürlek1, Ali Uyan1, Serpil Karan1, Servet Doğdu1 1Molecular Ecology and Fisheries Genetics Laboratory, Marine Science Department, Faculty of Marine Science and Technology, Iskenderun Technical University, 31220 Iskenderun, Hatay, Turkey 2Department of Biology, Faculty of Arts and Science, Duzce University, Duzce, Turkey Abstract Scopthalmus rhombus is rarely occurred and restricted to marine and estuarine sites in the eastern Marmara Sea and western Black Sea coast of Turkey. S. rhombus is occasionally caught in low numbers and continuously decreased in abundance due to overfishing and habitat degradations. This species should be considered to be threatened for Turkish marine waters. This species might also be recorded in the IUCN Red List of Threatened Species as Near Threatened (NT). Keywords: Scopthalmus rhombus, Threatened species, Marmara Sea, Western Black Sea, Turkey. Article history: Received 19 January 2016, Accepted 01 February 2016, Available online 02 February 2016 Introduction Brill Scopthalmus rhombus (Linnaeus, 1758) is a flatfish species belong to the family of Scopthalmidae. There are two more species of the genus Scopthalmus as Scopthalmus maximus and Scopthalmus maeticus for European fisheries and aquaculture. These three species are closely related congeneric species (Pardo et al., 2005; Azevedo et al., 2008; Turan, 2007) which show a similar distributional range (Blanquer et al., 1992; Pardo et al., 2001). S. rhombus is a commercial species and distributed on the parts of the Mediterranean Sea and Black Sea to the northeast Atlantic. -

A Case Study of North Sea Plaice and Sole
EVOLUTIONARY EFFECTS OF FISHING AND IMPLICATIONS FOR SUSTAINABLE MANAGEMENT: A CASE STUDY OF NORTH SEA PLAICE AND SOLE Fabian M. Mollet Thesis committee Thesis supervisor: Prof. dr. A.D. Rijnsdorp Professor of sustainable fisheries Wageningen University Other members: Prof. dr. M. Heino (University of Bergen, Norway) Prof. dr. R.F. Hoekstra (Wageningen University) Prof. dr. J.M. Tinbergen (University of Groningen) Dr. H.W. van der Veer (Royal Netherlands Institute of Sea Research, Den Burg) This research was conducted under the auspices of the Graduate School of Wageningen Institute for Animal Sciences (WIAS). EVOLUTIONARY EFFECTS OF FISHING AND IMPLICATIONS FOR SUSTAINABLE MANAGEMENT: A CASE STUDY OF NORTH SEA PLAICE AND SOLE Fabian M. Mollet Thesis Submitted in fulfilment of the requirements for the degree of doctor at Wageningen University by the authority of the Rector Magnificus Prof. dr. M.J. Kropff, in the presence of the Thesis Committee appointed by the Academic Board to be defended in public on Friday 7 May 2010 at 11 a.m. in the Aula. Fabian M. Mollet Evolutionary effects of fishing and implications for sustainable management: a case study of North Sea plaice and sole, 204 pages Thesis, Wageningen University, Wageningen, NL (2010) With references, with summaries in English ISBN 978-90-8585-613-9 Summary Exploited resources might genetically evolve as a consequence of ex- ploitation by adapting their life history to the imposed mortality re- gime. Although evolution favors traits for survival and reproduction of the fittest, human-induced evolution might have negative consequences for the exploiter. In general, a shift towards lower growth rate, earlier maturation and increased reproductive investment might be expected from increased (unselective) mortality and these changes might lead to generally smaller exploited individuals.