Centre for Basic Sciences
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Venter Institute
Genomics:GTL Program Projects J. Craig Venter Institute 42 Estimation of the Minimal Mycoplasma Gene Set Using Global Transposon Mutagenesis and Comparative Genomics John I. Glass* ([email protected]), Nina Alperovich, Nacyra Assad-Garcia, Shibu Yooseph, Mahir Maruf, Carole Lartigue, Cynthia Pfannkoch, Clyde A. Hutchison III, Hamilton O. Smith, and J. Craig Venter J. Craig Venter Institute, Rockville, MD The Venter Institute aspires to make bacteria with specific metabolic capabilities encoded by artificial genomes. To achieve this we must develop technologies and strategies for creating bacterial cells from constituent parts of either biological or synthetic origin. Determining the minimal gene set needed for a functioning bacterial genome in a defined laboratory environment is a necessary step towards our goal. For our initial rationally designed cell we plan to synthesize a genome based on a mycoplasma blueprint (mycoplasma being the common name for the class Mollicutes). We chose this bacterial taxon because its members already have small, near minimal genomes that encode limited metabolic capacity and complexity. We took two approaches to determine what genes would need to be included in a truly minimal synthetic chromosome of a planned Mycoplasma laboratorium: determination of all the non-essential genes through random transposon mutagenesis of model mycoplasma species, Mycoplasma genitalium, and comparative genomics of a set of 15 mycoplasma genomes in order to identify genes common to all members of the taxon. Global transposon mutagenesis has been used to predict the essential gene set for a number of bac- teria. In Bacillus subtilis all but 271 of bacterium’s ~4100 genes could be knocked out. -

DNA: the Future Storage Device
Special Issue - 2017 International Journal of Engineering Research & Technology (IJERT) ISSN: 2278-0181 ICADEMS - 2017 Conference Proceedings DNA: The Future Storage Device Sakshi Jha1, Mr. Mahesh Kumar Malkani2 1Ganga Institute of Technology and Management, 2Department of Computer Science Engineering and CFIS, MDU, Rohtak GITAM, MDU, Rohtak Data Storage has been a great concern since the early ages. than current magnetic tape or hard drive due to its huge With the advent of time, data has been expanding and new density .Digital data universe has been growing storage devices have been satisfying the demand. Demand for exponentially. Trends in data storage are setting new data storage is growing on a faster pace .In order to meet this milestones. From floppy disk to tape drives and tape drives growing need for storage device, a shift from electronic media to cloud storage have been the new developments in the to molecular DNA has been made. DNA molecule is universal and fundamental data storage unit in biology. 1gram of DNA field of storage devices. Most of the world’s data is stored molecule can store 1 Zettabyte of data. A new encoding on magnetic tapes and optical disk drives. Despite this scheme that offers controllable redundancy, trading off improvement, storing Zetta bytes of data would still take reliability for density has also been developed. Finally, we millions of years and space. With the increasing technology highlight trends in biotechnology that indicate the impending and tech-literacy, there is a need for revolution in the arena practicality of DNA storage for much larger datasets. of archiving. -

Microbial Identification Framework for Risk Assessment
Microbial Identification Framework for Risk Assessment May 2017 Cat. No.: En14-317/2018E-PDF ISBN 978-0-660-24940-7 Information contained in this publication or product may be reproduced, in part or in whole, and by any means, for personal or public non-commercial purposes, without charge or further permission, unless otherwise specified. You are asked to: • Exercise due diligence in ensuring the accuracy of the materials reproduced; • Indicate both the complete title of the materials reproduced, as well as the author organization; and • Indicate that the reproduction is a copy of an official work that is published by the Government of Canada and that the reproduction has not been produced in affiliation with or with the endorsement of the Government of Canada. Commercial reproduction and distribution is prohibited except with written permission from the author. For more information, please contact Environment and Climate Change Canada’s Inquiry Centre at 1-800-668-6767 (in Canada only) or 819-997-2800 or email to [email protected]. © Her Majesty the Queen in Right of Canada, represented by the Minister of the Environment and Climate Change, 2016. Aussi disponible en français Microbial Identification Framework for Risk Assessment Page 2 of 98 Summary The New Substances Notification Regulations (Organisms) (the regulations) of the Canadian Environmental Protection Act, 1999 (CEPA) are organized according to organism type (micro- organisms and organisms other than micro-organisms) and by activity. The Microbial Identification Framework for Risk Assessment (MIFRA) provides guidance on the required information for identifying micro-organisms. This document is intended for those who deal with the technical aspects of information elements or information requirements of the regulations that pertain to identification of a notified micro-organism. -

Role of Protein Phosphorylation in Mycoplasma Pneumoniae
Pathogenicity of a minimal organism: Role of protein phosphorylation in Mycoplasma pneumoniae Dissertation zur Erlangung des mathematisch-naturwissenschaftlichen Doktorgrades „Doctor rerum naturalium“ der Georg-August-Universität Göttingen vorgelegt von Sebastian Schmidl aus Bad Hersfeld Göttingen 2010 Mitglieder des Betreuungsausschusses: Referent: Prof. Dr. Jörg Stülke Koreferent: PD Dr. Michael Hoppert Tag der mündlichen Prüfung: 02.11.2010 “Everything should be made as simple as possible, but not simpler.” (Albert Einstein) Danksagung Zunächst möchte ich mich bei Prof. Dr. Jörg Stülke für die Ermöglichung dieser Doktorarbeit bedanken. Nicht zuletzt durch seine freundliche und engagierte Betreuung hat mir die Zeit viel Freude bereitet. Des Weiteren hat er mir alle Freiheiten zur Verwirklichung meiner eigenen Ideen gelassen, was ich sehr zu schätzen weiß. Für die Übernahme des Korreferates danke ich PD Dr. Michael Hoppert sowie Prof. Dr. Heinz Neumann, PD Dr. Boris Görke, PD Dr. Rolf Daniel und Prof. Dr. Botho Bowien für das Mitwirken im Thesis-Komitee. Der Studienstiftung des deutschen Volkes gilt ein besonderer Dank für die finanzielle Unterstützung dieser Arbeit, durch die es mir unter anderem auch möglich war, an Tagungen in fernen Ländern teilzunehmen. Prof. Dr. Michael Hecker und der Gruppe von Dr. Dörte Becher (Universität Greifswald) danke ich für die freundliche Zusammenarbeit bei der Durchführung von zahlreichen Proteomics-Experimenten. Ein ganz besonderer Dank geht dabei an Katrin Gronau, die mich in die Feinheiten der 2D-Gelelektrophorese eingeführt hat. Außerdem möchte ich mich bei Andreas Otto für die zahlreichen Proteinidentifikationen in den letzten Monaten bedanken. Nicht zu vergessen ist auch meine zweite Außenstelle an der Universität in Barcelona. Dr. Maria Lluch-Senar und Dr. -
Downloaded” to a Computer Than to Answer Questions About Emotions, Which Will Organize Their World
Between an Animal and a Machine MODERNITY IN QUESTION STUDIES IN PHILOSOPHY AND HISTORY OF IDEAS Edited by Małgorzata Kowalska VOLUME 10 Paweł Majewski Between an Animal and a Machine Stanisław Lem’s Technological Utopia Translation from Polish by Olga Kaczmarek Bibliographic Information published by the Deutsche Nationalbibliothek The Deutsche Nationalbibliothek lists this publication in the Deutsche Nationalbibliografie; detailed bibliographic data is available in the internet at http://dnb.d-nb.de. Library of Congress Cataloging-in-Publication Data A CIP catalog record for this book has been applied for at the Library of Congress. The Publication is founded by Ministry of Science and Higher Education of the Republic of Poland as a part of the National Programme for the Development of the Humanities. This publication reflects the views only of the authors, and the Ministry cannot be held responsible for any use which may be made of the information contained therein. ISSN 2193-3421 E-ISBN 978-3-653-06830-6 (E-PDF) E-ISBN 978-3-631-71024-1 (EPUB) E-ISBN 978-3-631-71025-8 (MOBI) DOI 10.3726/978-3-653-06830-6 Open Access: This work is licensed under a Creative Commons Attribution Non Commercial No Derivatives 4.0 unported license. To view a copy of this license, visit https://creativecommons.org/licenses/by-nc-nd/4.0/ © Paweł Majewski, 2018 . Peter Lang – Berlin · Bern · Bruxelles · New York · Oxford · Warszawa · Wien This publication has been peer reviewed. www.peterlang.com Contents Introduction ........................................................................................................ 9 Lemology Pure and Applied ............................................................................. 9 Part One Dialogues – Cybernetics as an Anthropology ........................................ -
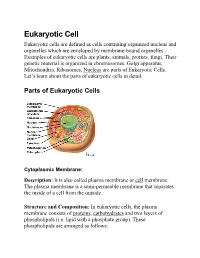
Eukaryotic Cell Eukaryotic Cells Are Defined As Cells Containing Organized Nucleus and Organelles Which Are Enveloped by Membrane-Bound Organelles
Eukaryotic Cell Eukaryotic cells are defined as cells containing organized nucleus and organelles which are enveloped by membrane-bound organelles. Examples of eukaryotic cells are plants, animals, protists, fungi. Their genetic material is organized in chromosomes. Golgi apparatus, Mitochondria, Ribosomes, Nucleus are parts of Eukaryotic Cells. Let’s learn about the parts of eukaryotic cells in detail. Parts ot Eukaryotic Cells Cytoplasmic Membrane: Description: It is also called plasma membrane or cell membrane. The plasma membrane is a semi-permeable membrane that separates the inside of a cell from the outside. Structure and Composition: In eukaryotic cells, the plasma membrane consists of proteins , carbohydrates and two layers of phospholipids (i.e. lipid with a phosphate group). These phospholipids are arranged as follows: • The polar, hydrophilic (water-loving) heads face the outside and inside of the cell. These heads interact with the aqueous environment outside and within a cell. • The non-polar, hydrophobic (water-repelling) tails are sandwiched between the heads and are protected from the aqueous environments. Scientists Singer and Nicolson(1972) described the structure of the phospholipid bilayer as the ‘Fluid Mosaic Model’. The reason is that the bi-layer looks like a mosaic and has a semi-fluid nature that allows lateral movement of proteins within the bilayer. Image: Fluid mosaic model. Orange circles – Hydrophilic heads; Lines below – Hydrophobic tails. Functions • The plasma membrane is selectively permeable i.e. it allows only selected substances to pass through. • It protects the cells from shock and injuries. • The fluid nature of the membrane allows the interaction of molecules within the membrane. -

Diagnostic Strategies for COVID-19 and Other Coronaviruses Medical Virology: from Pathogenesis to Disease Control
Medical Virology: From Pathogenesis to Disease Control Series Editor: Shailendra K. Saxena Pranjal Chandra Sharmili Roy Editors Diagnostic Strategies for COVID-19 and other Coronaviruses Medical Virology: From Pathogenesis to Disease Control Series Editor Shailendra K. Saxena, Centre for Advanced Research, King George’s Medical University, Lucknow, India This book series reviews the recent advancement in the field of medical virology including molecular epidemiology, diagnostics and therapeutic strategies for various viral infections. The individual books in this series provide a comprehensive over- view of infectious diseases that are caused by emerging and re-emerging viruses including their mode of infections, immunopathology, diagnosis, treatment, epide- miology, and etiology. It also discusses the clinical recommendations in the man- agement of infectious diseases focusing on the current practices, recent advances in diagnostic approaches and therapeutic strategies. The books also discuss progress and challenges in the development of viral vaccines and discuss the application of viruses in the translational research and human healthcare. More information about this series at http://www.springer.com/series/16573 Pranjal Chandra • Sharmili Roy Editors Diagnostic Strategies for COVID-19 and other Coronaviruses Editors Pranjal Chandra Sharmili Roy School of Biochemical Engineering Division of Oncology Indian Institute of Technology Stanford Medicine Varanasi, India California, CA, USA ISSN 2662-981X ISSN 2662-9828 (electronic) Medical Virology: From Pathogenesis to Disease Control ISBN 978-981-15-6005-7 ISBN 978-981-15-6006-4 (eBook) https://doi.org/10.1007/978-981-15-6006-4 # The Editor(s) (if applicable) and The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd. -

Appendix 1*) for Essential Information Very Well Illustrated in Google and Wikipedia
Appendix 1*) For Essential Information Very Well Illustrated in Google and Wikipedia *) in Support of the Text with Literature Citations. Referrals to illustrations in Appendix 2. Cancer in the Plant. The insertion of the Agrobacterium tumefaciens circular plasmid T (transferred) DNA into the genome of its new host, the plant (Gelvin BS. Microbiol Molecular Biol Rev 2003;67:16–37). The plant cancer “crown gall” (agrocallus; Agrobacterial crown gall) consists of malignantly transformed cells replicating the agrobacterial T DNA plasmid (reviewed in postscript Table XXXV). For reference: Koncz C Mayerhofer R Koncz-Kálmán Zs et al EMBO J 1990;9:1337–1346. Transfer of potentially oncogenic bacterial genes and proteins to patients: Septicemic Bacteroides enterotoxigenic (Sinkovics J G & Smith JP Cancer 1970;25:663–671; Viljoen KS et al PLoS One 2015;10(3):e0119462); Bartonella bacilliformis etc (Guy L et al PLoS Genet 2013;9(3):e1003393; Harms A & Dehio C Clin Microbiol Rev 2012;25:42–78; Llosa M et al Trends Microbiol 2012;20:355–9; Minnick MF et al PLoS Negl Trop Dis 2014;6(7):e2919); Helicobacter pylori (Bonsor DA et al J Biol Chem 2015;pii:jbc.M115.641829; Su YL et al J Immunol 2015;194:3997–4007; Vaziri F et al Pathog Dis 2015;73(3). pii.ftu021); Porphyromonas gingivalis (Katz J et al Int J Oral Sci 2011;3:209–215); Tuberculous infections with A. tumefaciens in patients (Ramirez FC et al Clin Infect Dis 1992;15:938–940). DNA-binding Antibodies. DNA- (or RNA-) binding proteins use zink finger motifs, leucine zippers and winged (beta-sheet loops) helix-turn helix motifs (HTH, two helices separated by the loop, RNA/DNA-binding domains) in recognition of RNA/DNA receptors for attachment. -

Coding of Still Pictures
ISO/IEC JTC 1/SC29/WG1N91016 91st Meeting, Online, 19-23 April 2021 ISO/IEC JTC 1/SC 29/WG 1 (ITU-T SG16) Coding of Still Pictures JBIG JPEG Joint Bi-level Image Joint Photographic Experts Group Experts Group TITLE: DNA-based Media Storage: State-of-the-Art, Challenges, Use Cases and Requirements version 4.0 SOURCE: Contributors: Marc Antonini, Luis Cruz, Eduardo da Silva, Melpomeni Dimopoulou, Touradj Ebrahimi (editor), Siegfried Foessel, Gloria Menegaz, Fernando Pereira (editor), António Pinheiro, Mohamad Raad PROJECT: JPEG DNA Exploration STATUS: Final REQUESTED ACTION: For information and feedback DISTRIBUTION: Public Contact: ISO/IEC JTC 1/SC 29/WG 1 Convener – Prof. Touradj Ebrahimi EPFL/STI/IEL/GR-EB, Station 11, CH-1015 Lausanne, Switzerland Tel: +41 21 693 2606, Fax: +41 21 693 7600, E-mail: [email protected] - 1 - ISO/IEC JTC 1/SC29/WG1N91016 91st Meeting, Online, 19-23 April 2021 DNA-based Media Storage State-of-the-Art, Challenges, Use Cases and Requirements Version 4.0 Executive Summary DNA is a macromolecule which is essential for any form of life and is made of simple units that line up in a particular order within this large molecule. The order of these units usually carries genetic information for a specific life organism, similar to how the order of letters in a text carries information. In practice, this means that DNA molecules may be artificially created with specific unit orders, notably to store some relevant sequence of information. In digital media information, notably images, the relevant representation symbols, e.g. quantized DCT coefficients, are expressed in bits (binary units) but they could be expressed in any other units, for example the DNA units which follow a quaternary (4-ary) representation basis. -

Nucleic Acid/Inorganic Particle Hybrid Systems from Design to Application
Research Collection Doctoral Thesis Nucleic acid/inorganic particle hybrid systems from design to application Author(s): Puddu, Michela Publication Date: 2015 Permanent Link: https://doi.org/10.3929/ethz-a-010609177 Rights / License: In Copyright - Non-Commercial Use Permitted This page was generated automatically upon download from the ETH Zurich Research Collection. For more information please consult the Terms of use. ETH Library DISS. ETH No. 23177 NUCLEIC ACID/INORGANIC PARTICLE HYBRID SYSTEMS: FROM DESIGN TO APPLICATION A thesis submitted to attain the degree of DOCTOR OF SCIENCES of ETH ZURICH (Dr. sc. ETH Zurich) presented by MICHELA PUDDU MSc Materials Science born on 02.03.1987 citizen of Italy accepted on the recommendation of Prof. Dr. Wendelin J. Stark, examiner Prof. Dr. János Vörös, co-examiner Dr. Robert N. Grass, co-examiner 2015 2 3 To my parents “Considerate la vostra semenza: fatti non foste a viver come bruti, ma per seguir virtute e canoscenza." Dante Alighieri, Divina Commedia, Inferno, canto XXVI, vv. 18-20. 4 5 Acknowledgments The PhD has been a wonderful and exciting experience, and I want to take a moment to thank the many people who took part in this journey. Foremost, I would like to express my gratitude to Prof. Wendelin Stark for giving me the opportunity to do research and develop a mature scientific approach in a highly stimulating and innovative environment. I am indebted to him for providing me with entrepreneurial spirit and vision that influenced my thinking and career choice. I also thank him for the precious lesson that a balance between research interests and personal pursuits does not keep away success, but it actually brings one step closer to it. -

AST 201 - Introduction to Astrobiology Script
AST 201 - Introduction to Astrobiology Script Samuel Gunz1 and Martin Emons1 1Department of Biosystems Science and Engineering, ETH Zurich Autumn 2020 1 What is Life? Note that also some non-living things satisfy some of these traits (e.g. Fire, snowflake). In addition, some living things The definition of Life is not simple. NASA defines life as (e.g. seed, bacteria) can undergo a period of dormancy. Are follows, “Life is a self-sustaining system capable of Dar- they dead during that time because they are not growing, winian evolution.”. However, this excludes for instance metabolising or interacting with the environment? mules as they are infertile. The definition might depend in which context it is asked. Different smart people came up with various definitions, however they could never agree 1.2 Physical definition of life upon a single definition. It is important that a definition Life is an ordered system of molecules that ‘disobeys’ the should apply to alien life as well. Reproduction/replication second law of thermodynamics – that entropy always in- needs to be imperfect to allow for natural selection of those creases. An isolated cell cannot violate the second law branches of life with beneficial traits, otherwise life would of thermodynamics, the only way it can maintain a low- have got stuck at the first replicating organism. entropy, nonequilibrium state characterised by a high de- gree of structural organisation is to increase the entropy of The chicken and egg problem illustrates the problem of its surroundings. A cell releases some of the energy that it the definition of life and species. -

M.Sc. Microbiology (2019 ONWARDS)
PONDICHERRY UNIVERSITY PUDUCHERRY 605 014 CURRICULUM AND SYLLABUS of M.Sc. Microbiology (2019 ONWARDS) Department of Microbiology School of Life Sciences About the course The Department of Microbiology is committed to excellence in education, research and extension. This Department is being strengthened with various research units and periodical update / modernization of the curricula. The Department of Microbiology at the Pondicherry University, School of Life Sciences, brings together a variety of researchers as faculty of this programme who are specialized in their domains and united by the common goal of understanding the “Microbes”. Microbes are playing important role in the bioprocess of all living things and maintain homeostasis of the universe. Without microbes, one cannot imagine such a biologically balanced and diverse universe; rather our earth would have placed as a barren planet. As the microbial activities are so diverse, the microbiology programme is a multidisciplinary subject, which will have the roots of life science, environmental science, and engineering. Traditional microbiology is considered to be an important area of study in biology since it has enormous potential and vast scope in fermentation, bioremediation and biomedical technology. But the recent developments from human microbiome project, metagenomics and microbial genome projects has expanded its scope and potential in the next generation drug design, molecular pathogenesis, phylogeography, production of smart biomolecules, etc. Modern Microbiology has expanded its roots in genome technology, nanobiotechnology, green energy (biofuel) technology, bioelectronics etc. Considering recent innovations and rapid growth of microbiological approaches and applications in human and environmental sustainability, the M.Sc. Microbiology curricula is designed to enlighten the students in basics of Microbiology to recent developments.