GTL PI Meeting Abstracts
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Venter Institute
Genomics:GTL Program Projects J. Craig Venter Institute 42 Estimation of the Minimal Mycoplasma Gene Set Using Global Transposon Mutagenesis and Comparative Genomics John I. Glass* ([email protected]), Nina Alperovich, Nacyra Assad-Garcia, Shibu Yooseph, Mahir Maruf, Carole Lartigue, Cynthia Pfannkoch, Clyde A. Hutchison III, Hamilton O. Smith, and J. Craig Venter J. Craig Venter Institute, Rockville, MD The Venter Institute aspires to make bacteria with specific metabolic capabilities encoded by artificial genomes. To achieve this we must develop technologies and strategies for creating bacterial cells from constituent parts of either biological or synthetic origin. Determining the minimal gene set needed for a functioning bacterial genome in a defined laboratory environment is a necessary step towards our goal. For our initial rationally designed cell we plan to synthesize a genome based on a mycoplasma blueprint (mycoplasma being the common name for the class Mollicutes). We chose this bacterial taxon because its members already have small, near minimal genomes that encode limited metabolic capacity and complexity. We took two approaches to determine what genes would need to be included in a truly minimal synthetic chromosome of a planned Mycoplasma laboratorium: determination of all the non-essential genes through random transposon mutagenesis of model mycoplasma species, Mycoplasma genitalium, and comparative genomics of a set of 15 mycoplasma genomes in order to identify genes common to all members of the taxon. Global transposon mutagenesis has been used to predict the essential gene set for a number of bac- teria. In Bacillus subtilis all but 271 of bacterium’s ~4100 genes could be knocked out. -

Characterization of Stress Tolerance and Metabolic Capabilities of Acidophilic Iron-Sulfur-Transforming Bacteria and Their Relevance to Mars
Characterization of stress tolerance and metabolic capabilities of acidophilic iron-sulfur-transforming bacteria and their relevance to Mars Dissertation zur Erlangung des akademischen Grades eines Doktors der Naturwissenschaften – Dr. rer. nat. – vorgelegt von Anja Bauermeister aus Leipzig Im Fachbereich Chemie der Universität Duisburg-Essen 2012 Die vorliegende Arbeit wurde im Zeitraum von März 2009 bis Dezember 2012 im Arbeitskreis von Prof. Dr. Hans-Curt Flemming am Biofilm Centre (Fakultät für Chemie) der Universität Duisburg-Essen und in der Abteilung Strahlenbiologie (Institut für Luft- und Raumfahrtmedizin, Deutsches Zentrum für Luft- und Raumfahrt, Köln) durchgeführt. Tag der Einreichung: 07.12.2012 Tag der Disputation: 23.04.2013 Gutachter: Prof. Dr. H.-C. Flemming Prof. Dr. W. Sand Vorsitzender: Prof. Dr. C. Mayer Erklärung / Statement Hiermit versichere ich, dass ich die vorliegende Arbeit mit dem Titel „Characterization of stress tolerance and metabolic capabilities of acidophilic iron- sulfur-transforming bacteria and their relevance to Mars” selbst verfasst und keine außer den angegebenen Hilfsmitteln und Quellen benutzt habe, und dass die Arbeit in dieser oder ähnlicher Form noch bei keiner anderen Universität eingereicht wurde. Herewith I declare that this thesis is the result of my independent work. All sources and auxiliary materials used by me in this thesis are cited completely. Essen, 07.12.2012 Table of contents Abbreviations ....................................................................................... -

Microbial Identification Framework for Risk Assessment
Microbial Identification Framework for Risk Assessment May 2017 Cat. No.: En14-317/2018E-PDF ISBN 978-0-660-24940-7 Information contained in this publication or product may be reproduced, in part or in whole, and by any means, for personal or public non-commercial purposes, without charge or further permission, unless otherwise specified. You are asked to: • Exercise due diligence in ensuring the accuracy of the materials reproduced; • Indicate both the complete title of the materials reproduced, as well as the author organization; and • Indicate that the reproduction is a copy of an official work that is published by the Government of Canada and that the reproduction has not been produced in affiliation with or with the endorsement of the Government of Canada. Commercial reproduction and distribution is prohibited except with written permission from the author. For more information, please contact Environment and Climate Change Canada’s Inquiry Centre at 1-800-668-6767 (in Canada only) or 819-997-2800 or email to [email protected]. © Her Majesty the Queen in Right of Canada, represented by the Minister of the Environment and Climate Change, 2016. Aussi disponible en français Microbial Identification Framework for Risk Assessment Page 2 of 98 Summary The New Substances Notification Regulations (Organisms) (the regulations) of the Canadian Environmental Protection Act, 1999 (CEPA) are organized according to organism type (micro- organisms and organisms other than micro-organisms) and by activity. The Microbial Identification Framework for Risk Assessment (MIFRA) provides guidance on the required information for identifying micro-organisms. This document is intended for those who deal with the technical aspects of information elements or information requirements of the regulations that pertain to identification of a notified micro-organism. -

The Effects of Natural Variation in Background Radioactivity on Humans, Animals and Other Organisms
Biol. Rev. (2012), pp. 000–000. 1 doi: 10.1111/j.1469-185X.2012.00249.x The effects of natural variation in background radioactivity on humans, animals and other organisms Anders P. Møller1,∗ and Timothy A. Mousseau2 1Laboratoire d’Ecologie, Syst´ematique et Evolution, CNRS UMR 8079, Universit´e Paris-Sud, Bˆatiment 362, F-91405, Orsay Cedex, France 2Department of Biological Sciences, University of South Carolina, Columbia, SC 29208, USA ABSTRACT Natural levels of radioactivity on the Earth vary by more than a thousand-fold; this spatial heterogeneity may suffice to create heterogeneous effects on physiology, mutation and selection. We review the literature on the relationship between variation in natural levels of radioactivity and evolution. First, we consider the effects of natural levels of radiation on mutations, DNA repair and genetics. A total of 46 studies with 373 effect size estimates revealed a small, but highly significant mean effect that was independent of adjustment for publication bias. Second, we found different mean effect sizes when studies were based on broad categories like physiology, immunology and disease frequency; mean weighted effect sizes were larger for studies of plants than animals, and larger in studies conducted in areas with higher levels of radiation. Third, these negative effects of radiation on mutations, immunology and life history are inconsistent with a general role of hormetic positive effects of radiation on living organisms. Fourth, we reviewed studies of radiation resistance among taxa. These studies suggest that current levels of natural radioactivity may affect mutational input and thereby the genetic constitution and composition of natural populations. -

Role of Protein Phosphorylation in Mycoplasma Pneumoniae
Pathogenicity of a minimal organism: Role of protein phosphorylation in Mycoplasma pneumoniae Dissertation zur Erlangung des mathematisch-naturwissenschaftlichen Doktorgrades „Doctor rerum naturalium“ der Georg-August-Universität Göttingen vorgelegt von Sebastian Schmidl aus Bad Hersfeld Göttingen 2010 Mitglieder des Betreuungsausschusses: Referent: Prof. Dr. Jörg Stülke Koreferent: PD Dr. Michael Hoppert Tag der mündlichen Prüfung: 02.11.2010 “Everything should be made as simple as possible, but not simpler.” (Albert Einstein) Danksagung Zunächst möchte ich mich bei Prof. Dr. Jörg Stülke für die Ermöglichung dieser Doktorarbeit bedanken. Nicht zuletzt durch seine freundliche und engagierte Betreuung hat mir die Zeit viel Freude bereitet. Des Weiteren hat er mir alle Freiheiten zur Verwirklichung meiner eigenen Ideen gelassen, was ich sehr zu schätzen weiß. Für die Übernahme des Korreferates danke ich PD Dr. Michael Hoppert sowie Prof. Dr. Heinz Neumann, PD Dr. Boris Görke, PD Dr. Rolf Daniel und Prof. Dr. Botho Bowien für das Mitwirken im Thesis-Komitee. Der Studienstiftung des deutschen Volkes gilt ein besonderer Dank für die finanzielle Unterstützung dieser Arbeit, durch die es mir unter anderem auch möglich war, an Tagungen in fernen Ländern teilzunehmen. Prof. Dr. Michael Hecker und der Gruppe von Dr. Dörte Becher (Universität Greifswald) danke ich für die freundliche Zusammenarbeit bei der Durchführung von zahlreichen Proteomics-Experimenten. Ein ganz besonderer Dank geht dabei an Katrin Gronau, die mich in die Feinheiten der 2D-Gelelektrophorese eingeführt hat. Außerdem möchte ich mich bei Andreas Otto für die zahlreichen Proteinidentifikationen in den letzten Monaten bedanken. Nicht zu vergessen ist auch meine zweite Außenstelle an der Universität in Barcelona. Dr. Maria Lluch-Senar und Dr. -

Deinococcus Antarcticus Sp. Nov., Isolated from Soil
International Journal of Systematic and Evolutionary Microbiology (2015), 65, 331–335 DOI 10.1099/ijs.0.066324-0 Deinococcus antarcticus sp. nov., isolated from soil Ning Dong,1,2 Hui-Rong Li,1 Meng Yuan,1,2 Xiao-Hua Zhang2 and Yong Yu1 Correspondence 1SOA Key Laboratory for Polar Science, Polar Research Institute of China, Shanghai 200136, Yong Yu PR China [email protected] 2College of Marine Life Sciences, Ocean University of China, Qingdao 266003, PR China A pink-pigmented, non-motile, coccoid bacterial strain, designated G3-6-20T, was isolated from a soil sample collected in the Grove Mountains, East Antarctica. This strain was resistant to UV irradiation (810 J m”2) and slightly more sensitive to desiccation as compared with Deinococcus radiodurans. Phylogenetic analyses based on the 16S rRNA gene sequence of the isolate indicated that the organism belongs to the genus Deinococcus. Highest sequence similarities were with Deinococcus ficus CC-FR2-10T (93.5 %), Deinococcus xinjiangensis X-82T (92.8 %), Deinococcus indicus Wt/1aT (92.5 %), Deinococcus daejeonensis MJ27T (92.3 %), Deinococcus wulumuqiensis R-12T (92.3 %), Deinococcus aquaticus PB314T (92.2 %) and T Deinococcus radiodurans DSM 20539 (92.2 %). Major fatty acids were C18 : 1v7c, summed feature 3 (C16 : 1v7c and/or C16 : 1v6c), anteiso-C15 : 0 and C16 : 0. The G+C content of the genomic DNA of strain G3-6-20T was 63.1 mol%. Menaquinone 8 (MK-8) was the predominant respiratory quinone. Based on its phylogenetic position, and chemotaxonomic and phenotypic characteristics, strain G3-6-20T represents a novel species of the genus Deinococcus, for which the name Deinococcus antarcticus sp. -

Access to Electronic Thesis
Access to Electronic Thesis Author: Khalid Salim Al-Abri Thesis title: USE OF MOLECULAR APPROACHES TO STUDY THE OCCURRENCE OF EXTREMOPHILES AND EXTREMODURES IN NON-EXTREME ENVIRONMENTS Qualification: PhD This electronic thesis is protected by the Copyright, Designs and Patents Act 1988. No reproduction is permitted without consent of the author. It is also protected by the Creative Commons Licence allowing Attributions-Non-commercial-No derivatives. If this electronic thesis has been edited by the author it will be indicated as such on the title page and in the text. USE OF MOLECULAR APPROACHES TO STUDY THE OCCURRENCE OF EXTREMOPHILES AND EXTREMODURES IN NON-EXTREME ENVIRONMENTS By Khalid Salim Al-Abri Msc., University of Sultan Qaboos, Muscat, Oman Mphil, University of Sheffield, England Thesis submitted in partial fulfillment for the requirements of the Degree of Doctor of Philosophy in the Department of Molecular Biology and Biotechnology, University of Sheffield, England 2011 Introductory Pages I DEDICATION To the memory of my father, loving mother, wife “Muneera” and son “Anas”, brothers and sisters. Introductory Pages II ACKNOWLEDGEMENTS Above all, I thank Allah for helping me in completing this project. I wish to express my thanks to my supervisor Professor Milton Wainwright, for his guidance, supervision, support, understanding and help in this project. In addition, he also stood beside me in all difficulties that faced me during study. My thanks are due to Dr. D. J. Gilmour for his co-supervision, technical assistance, his time and understanding that made some of my laboratory work easier. In the Ministry of Regional Municipalities and Water Resources, I am particularly grateful to Engineer Said Al Alawi, Director General of Health Control, for allowing me to carry out my PhD study at the University of Sheffield. -
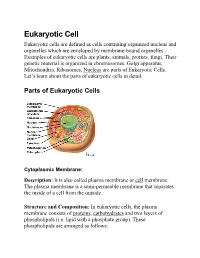
Eukaryotic Cell Eukaryotic Cells Are Defined As Cells Containing Organized Nucleus and Organelles Which Are Enveloped by Membrane-Bound Organelles
Eukaryotic Cell Eukaryotic cells are defined as cells containing organized nucleus and organelles which are enveloped by membrane-bound organelles. Examples of eukaryotic cells are plants, animals, protists, fungi. Their genetic material is organized in chromosomes. Golgi apparatus, Mitochondria, Ribosomes, Nucleus are parts of Eukaryotic Cells. Let’s learn about the parts of eukaryotic cells in detail. Parts ot Eukaryotic Cells Cytoplasmic Membrane: Description: It is also called plasma membrane or cell membrane. The plasma membrane is a semi-permeable membrane that separates the inside of a cell from the outside. Structure and Composition: In eukaryotic cells, the plasma membrane consists of proteins , carbohydrates and two layers of phospholipids (i.e. lipid with a phosphate group). These phospholipids are arranged as follows: • The polar, hydrophilic (water-loving) heads face the outside and inside of the cell. These heads interact with the aqueous environment outside and within a cell. • The non-polar, hydrophobic (water-repelling) tails are sandwiched between the heads and are protected from the aqueous environments. Scientists Singer and Nicolson(1972) described the structure of the phospholipid bilayer as the ‘Fluid Mosaic Model’. The reason is that the bi-layer looks like a mosaic and has a semi-fluid nature that allows lateral movement of proteins within the bilayer. Image: Fluid mosaic model. Orange circles – Hydrophilic heads; Lines below – Hydrophobic tails. Functions • The plasma membrane is selectively permeable i.e. it allows only selected substances to pass through. • It protects the cells from shock and injuries. • The fluid nature of the membrane allows the interaction of molecules within the membrane. -

Appendices Physico-Chemical
http://researchcommons.waikato.ac.nz/ Research Commons at the University of Waikato Copyright Statement: The digital copy of this thesis is protected by the Copyright Act 1994 (New Zealand). The thesis may be consulted by you, provided you comply with the provisions of the Act and the following conditions of use: Any use you make of these documents or images must be for research or private study purposes only, and you may not make them available to any other person. Authors control the copyright of their thesis. You will recognise the author’s right to be identified as the author of the thesis, and due acknowledgement will be made to the author where appropriate. You will obtain the author’s permission before publishing any material from the thesis. An Investigation of Microbial Communities Across Two Extreme Geothermal Gradients on Mt. Erebus, Victoria Land, Antarctica A thesis submitted in partial fulfilment of the requirements for the degree of Master’s Degree of Science at The University of Waikato by Emily Smith Year of submission 2021 Abstract The geothermal fumaroles present on Mt. Erebus, Antarctica, are home to numerous unique and possibly endemic bacteria. The isolated nature of Mt. Erebus provides an opportunity to closely examine how geothermal physico-chemistry drives microbial community composition and structure. This study aimed at determining the effect of physico-chemical drivers on microbial community composition and structure along extreme thermal and geochemical gradients at two sites on Mt. Erebus: Tramway Ridge and Western Crater. Microbial community structure and physico-chemical soil characteristics were assessed via metabarcoding (16S rRNA) and geochemistry (temperature, pH, total carbon (TC), total nitrogen (TN) and ICP-MS elemental analysis along a thermal gradient 10 °C–64 °C), which also defined a geochemical gradient. -

Appendix 1*) for Essential Information Very Well Illustrated in Google and Wikipedia
Appendix 1*) For Essential Information Very Well Illustrated in Google and Wikipedia *) in Support of the Text with Literature Citations. Referrals to illustrations in Appendix 2. Cancer in the Plant. The insertion of the Agrobacterium tumefaciens circular plasmid T (transferred) DNA into the genome of its new host, the plant (Gelvin BS. Microbiol Molecular Biol Rev 2003;67:16–37). The plant cancer “crown gall” (agrocallus; Agrobacterial crown gall) consists of malignantly transformed cells replicating the agrobacterial T DNA plasmid (reviewed in postscript Table XXXV). For reference: Koncz C Mayerhofer R Koncz-Kálmán Zs et al EMBO J 1990;9:1337–1346. Transfer of potentially oncogenic bacterial genes and proteins to patients: Septicemic Bacteroides enterotoxigenic (Sinkovics J G & Smith JP Cancer 1970;25:663–671; Viljoen KS et al PLoS One 2015;10(3):e0119462); Bartonella bacilliformis etc (Guy L et al PLoS Genet 2013;9(3):e1003393; Harms A & Dehio C Clin Microbiol Rev 2012;25:42–78; Llosa M et al Trends Microbiol 2012;20:355–9; Minnick MF et al PLoS Negl Trop Dis 2014;6(7):e2919); Helicobacter pylori (Bonsor DA et al J Biol Chem 2015;pii:jbc.M115.641829; Su YL et al J Immunol 2015;194:3997–4007; Vaziri F et al Pathog Dis 2015;73(3). pii.ftu021); Porphyromonas gingivalis (Katz J et al Int J Oral Sci 2011;3:209–215); Tuberculous infections with A. tumefaciens in patients (Ramirez FC et al Clin Infect Dis 1992;15:938–940). DNA-binding Antibodies. DNA- (or RNA-) binding proteins use zink finger motifs, leucine zippers and winged (beta-sheet loops) helix-turn helix motifs (HTH, two helices separated by the loop, RNA/DNA-binding domains) in recognition of RNA/DNA receptors for attachment. -

AST 201 - Introduction to Astrobiology Script
AST 201 - Introduction to Astrobiology Script Samuel Gunz1 and Martin Emons1 1Department of Biosystems Science and Engineering, ETH Zurich Autumn 2020 1 What is Life? Note that also some non-living things satisfy some of these traits (e.g. Fire, snowflake). In addition, some living things The definition of Life is not simple. NASA defines life as (e.g. seed, bacteria) can undergo a period of dormancy. Are follows, “Life is a self-sustaining system capable of Dar- they dead during that time because they are not growing, winian evolution.”. However, this excludes for instance metabolising or interacting with the environment? mules as they are infertile. The definition might depend in which context it is asked. Different smart people came up with various definitions, however they could never agree 1.2 Physical definition of life upon a single definition. It is important that a definition Life is an ordered system of molecules that ‘disobeys’ the should apply to alien life as well. Reproduction/replication second law of thermodynamics – that entropy always in- needs to be imperfect to allow for natural selection of those creases. An isolated cell cannot violate the second law branches of life with beneficial traits, otherwise life would of thermodynamics, the only way it can maintain a low- have got stuck at the first replicating organism. entropy, nonequilibrium state characterised by a high de- gree of structural organisation is to increase the entropy of The chicken and egg problem illustrates the problem of its surroundings. A cell releases some of the energy that it the definition of life and species. -

M.Sc. Microbiology (2019 ONWARDS)
PONDICHERRY UNIVERSITY PUDUCHERRY 605 014 CURRICULUM AND SYLLABUS of M.Sc. Microbiology (2019 ONWARDS) Department of Microbiology School of Life Sciences About the course The Department of Microbiology is committed to excellence in education, research and extension. This Department is being strengthened with various research units and periodical update / modernization of the curricula. The Department of Microbiology at the Pondicherry University, School of Life Sciences, brings together a variety of researchers as faculty of this programme who are specialized in their domains and united by the common goal of understanding the “Microbes”. Microbes are playing important role in the bioprocess of all living things and maintain homeostasis of the universe. Without microbes, one cannot imagine such a biologically balanced and diverse universe; rather our earth would have placed as a barren planet. As the microbial activities are so diverse, the microbiology programme is a multidisciplinary subject, which will have the roots of life science, environmental science, and engineering. Traditional microbiology is considered to be an important area of study in biology since it has enormous potential and vast scope in fermentation, bioremediation and biomedical technology. But the recent developments from human microbiome project, metagenomics and microbial genome projects has expanded its scope and potential in the next generation drug design, molecular pathogenesis, phylogeography, production of smart biomolecules, etc. Modern Microbiology has expanded its roots in genome technology, nanobiotechnology, green energy (biofuel) technology, bioelectronics etc. Considering recent innovations and rapid growth of microbiological approaches and applications in human and environmental sustainability, the M.Sc. Microbiology curricula is designed to enlighten the students in basics of Microbiology to recent developments.