AST 201 - Introduction to Astrobiology Script
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Venter Institute
Genomics:GTL Program Projects J. Craig Venter Institute 42 Estimation of the Minimal Mycoplasma Gene Set Using Global Transposon Mutagenesis and Comparative Genomics John I. Glass* ([email protected]), Nina Alperovich, Nacyra Assad-Garcia, Shibu Yooseph, Mahir Maruf, Carole Lartigue, Cynthia Pfannkoch, Clyde A. Hutchison III, Hamilton O. Smith, and J. Craig Venter J. Craig Venter Institute, Rockville, MD The Venter Institute aspires to make bacteria with specific metabolic capabilities encoded by artificial genomes. To achieve this we must develop technologies and strategies for creating bacterial cells from constituent parts of either biological or synthetic origin. Determining the minimal gene set needed for a functioning bacterial genome in a defined laboratory environment is a necessary step towards our goal. For our initial rationally designed cell we plan to synthesize a genome based on a mycoplasma blueprint (mycoplasma being the common name for the class Mollicutes). We chose this bacterial taxon because its members already have small, near minimal genomes that encode limited metabolic capacity and complexity. We took two approaches to determine what genes would need to be included in a truly minimal synthetic chromosome of a planned Mycoplasma laboratorium: determination of all the non-essential genes through random transposon mutagenesis of model mycoplasma species, Mycoplasma genitalium, and comparative genomics of a set of 15 mycoplasma genomes in order to identify genes common to all members of the taxon. Global transposon mutagenesis has been used to predict the essential gene set for a number of bac- teria. In Bacillus subtilis all but 271 of bacterium’s ~4100 genes could be knocked out. -

Microbial Identification Framework for Risk Assessment
Microbial Identification Framework for Risk Assessment May 2017 Cat. No.: En14-317/2018E-PDF ISBN 978-0-660-24940-7 Information contained in this publication or product may be reproduced, in part or in whole, and by any means, for personal or public non-commercial purposes, without charge or further permission, unless otherwise specified. You are asked to: • Exercise due diligence in ensuring the accuracy of the materials reproduced; • Indicate both the complete title of the materials reproduced, as well as the author organization; and • Indicate that the reproduction is a copy of an official work that is published by the Government of Canada and that the reproduction has not been produced in affiliation with or with the endorsement of the Government of Canada. Commercial reproduction and distribution is prohibited except with written permission from the author. For more information, please contact Environment and Climate Change Canada’s Inquiry Centre at 1-800-668-6767 (in Canada only) or 819-997-2800 or email to [email protected]. © Her Majesty the Queen in Right of Canada, represented by the Minister of the Environment and Climate Change, 2016. Aussi disponible en français Microbial Identification Framework for Risk Assessment Page 2 of 98 Summary The New Substances Notification Regulations (Organisms) (the regulations) of the Canadian Environmental Protection Act, 1999 (CEPA) are organized according to organism type (micro- organisms and organisms other than micro-organisms) and by activity. The Microbial Identification Framework for Risk Assessment (MIFRA) provides guidance on the required information for identifying micro-organisms. This document is intended for those who deal with the technical aspects of information elements or information requirements of the regulations that pertain to identification of a notified micro-organism. -

Role of Protein Phosphorylation in Mycoplasma Pneumoniae
Pathogenicity of a minimal organism: Role of protein phosphorylation in Mycoplasma pneumoniae Dissertation zur Erlangung des mathematisch-naturwissenschaftlichen Doktorgrades „Doctor rerum naturalium“ der Georg-August-Universität Göttingen vorgelegt von Sebastian Schmidl aus Bad Hersfeld Göttingen 2010 Mitglieder des Betreuungsausschusses: Referent: Prof. Dr. Jörg Stülke Koreferent: PD Dr. Michael Hoppert Tag der mündlichen Prüfung: 02.11.2010 “Everything should be made as simple as possible, but not simpler.” (Albert Einstein) Danksagung Zunächst möchte ich mich bei Prof. Dr. Jörg Stülke für die Ermöglichung dieser Doktorarbeit bedanken. Nicht zuletzt durch seine freundliche und engagierte Betreuung hat mir die Zeit viel Freude bereitet. Des Weiteren hat er mir alle Freiheiten zur Verwirklichung meiner eigenen Ideen gelassen, was ich sehr zu schätzen weiß. Für die Übernahme des Korreferates danke ich PD Dr. Michael Hoppert sowie Prof. Dr. Heinz Neumann, PD Dr. Boris Görke, PD Dr. Rolf Daniel und Prof. Dr. Botho Bowien für das Mitwirken im Thesis-Komitee. Der Studienstiftung des deutschen Volkes gilt ein besonderer Dank für die finanzielle Unterstützung dieser Arbeit, durch die es mir unter anderem auch möglich war, an Tagungen in fernen Ländern teilzunehmen. Prof. Dr. Michael Hecker und der Gruppe von Dr. Dörte Becher (Universität Greifswald) danke ich für die freundliche Zusammenarbeit bei der Durchführung von zahlreichen Proteomics-Experimenten. Ein ganz besonderer Dank geht dabei an Katrin Gronau, die mich in die Feinheiten der 2D-Gelelektrophorese eingeführt hat. Außerdem möchte ich mich bei Andreas Otto für die zahlreichen Proteinidentifikationen in den letzten Monaten bedanken. Nicht zu vergessen ist auch meine zweite Außenstelle an der Universität in Barcelona. Dr. Maria Lluch-Senar und Dr. -

State Party Response to the Supplimentary Information Requested by Iucn for the Barberton Makhonjwa Mountains World Heritage
STATE PARTY RESPONSE TO THE SUPPLIMENTARY INFORMATION REQUESTED BY IUCN FOR THE BARBERTON MAKHONJWA MOUNTAINS WORLD HERITAGE NOMINATION 21 FEBRUARY 2018 GOVERNMENT OF THE REPUBLIC OF SOUTH AFRICA Barberton Makhonjwa Mountains is a site that South Africa submitted to Unesco for inscription as a World Heritage Site. An IUCN Evaluation Mission was successfully undertaken in September 2017. The aim of this Evaluation Mission was for IUCN to evaluate whether or not the property has Outstanding Universal Value, meet the conditions of integrity and (where relevant) of authenticity and meet the requirements of protection and management. Following the Evaluation Mission, IUCN wrote a letter to South Africa dated 20 December 2017, requesting supplementary information before finalising its recommendations to the World Heritage Committee. South Africa was requested to address the following aspects and to submit responses by 28 February 2018: Page: 1. Global Comparative analysis; 3 2. Legal protection; 3 3. Mining; 3 4. Buffer zones; 4 5. Relocation of people; 5 6. Threats; 5 7. Private Landowners; 5 8. Transboundary collaboration. 6 What follows below are the responses by South Africa, addressing the above points in chronological order, together with relevant attachments where indicated. 2 1. Global Comparative analysis As proposed by IUCN in terms of the Global Comparative Analysis, further review as requested has been submitted by Christoph Heubeck1, Carl Anhaeusser2 and Dion Brandt3. This new addendum has been integrated with the existing comparative analysis to provide a consolidated statement of the comparative values of the nominated property relative to other sites globally provided within the updated Nomination Dossier. -

Geology of the Eoarchean, >3.95 Ga, Nulliak Supracrustal
ÔØ ÅÒÙ×Ö ÔØ Geology of the Eoarchean, > 3.95 Ga, Nulliak supracrustal rocks in the Saglek Block, northern Labrador, Canada: The oldest geological evidence for plate tectonics Tsuyoshi Komiya, Shinji Yamamoto, Shogo Aoki, Yusuke Sawaki, Akira Ishikawa, Takayuki Tashiro, Keiko Koshida, Masanori Shimojo, Kazumasa Aoki, Kenneth D. Collerson PII: S0040-1951(15)00269-3 DOI: doi: 10.1016/j.tecto.2015.05.003 Reference: TECTO 126618 To appear in: Tectonophysics Received date: 30 December 2014 Revised date: 30 April 2015 Accepted date: 17 May 2015 Please cite this article as: Komiya, Tsuyoshi, Yamamoto, Shinji, Aoki, Shogo, Sawaki, Yusuke, Ishikawa, Akira, Tashiro, Takayuki, Koshida, Keiko, Shimojo, Masanori, Aoki, Kazumasa, Collerson, Kenneth D., Geology of the Eoarchean, > 3.95 Ga, Nulliak supracrustal rocks in the Saglek Block, northern Labrador, Canada: The oldest geological evidence for plate tectonics, Tectonophysics (2015), doi: 10.1016/j.tecto.2015.05.003 This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. ACCEPTED MANUSCRIPT Geology of the Eoarchean, >3.95 Ga, Nulliak supracrustal rocks in the Saglek Block, northern Labrador, Canada: The oldest geological evidence for plate tectonics Tsuyoshi Komiya1*, Shinji Yamamoto1, Shogo Aoki1, Yusuke Sawaki2, Akira Ishikawa1, Takayuki Tashiro1, Keiko Koshida1, Masanori Shimojo1, Kazumasa Aoki1 and Kenneth D. -
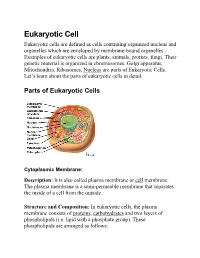
Eukaryotic Cell Eukaryotic Cells Are Defined As Cells Containing Organized Nucleus and Organelles Which Are Enveloped by Membrane-Bound Organelles
Eukaryotic Cell Eukaryotic cells are defined as cells containing organized nucleus and organelles which are enveloped by membrane-bound organelles. Examples of eukaryotic cells are plants, animals, protists, fungi. Their genetic material is organized in chromosomes. Golgi apparatus, Mitochondria, Ribosomes, Nucleus are parts of Eukaryotic Cells. Let’s learn about the parts of eukaryotic cells in detail. Parts ot Eukaryotic Cells Cytoplasmic Membrane: Description: It is also called plasma membrane or cell membrane. The plasma membrane is a semi-permeable membrane that separates the inside of a cell from the outside. Structure and Composition: In eukaryotic cells, the plasma membrane consists of proteins , carbohydrates and two layers of phospholipids (i.e. lipid with a phosphate group). These phospholipids are arranged as follows: • The polar, hydrophilic (water-loving) heads face the outside and inside of the cell. These heads interact with the aqueous environment outside and within a cell. • The non-polar, hydrophobic (water-repelling) tails are sandwiched between the heads and are protected from the aqueous environments. Scientists Singer and Nicolson(1972) described the structure of the phospholipid bilayer as the ‘Fluid Mosaic Model’. The reason is that the bi-layer looks like a mosaic and has a semi-fluid nature that allows lateral movement of proteins within the bilayer. Image: Fluid mosaic model. Orange circles – Hydrophilic heads; Lines below – Hydrophobic tails. Functions • The plasma membrane is selectively permeable i.e. it allows only selected substances to pass through. • It protects the cells from shock and injuries. • The fluid nature of the membrane allows the interaction of molecules within the membrane. -

Implications for Global-Scale Plate Tectonics Hamed Gamal El Dien1,2 ✉ , Luc S
www.nature.com/scientificreports OPEN Geochemical evidence for a widespread mantle re-enrichment 3.2 billion years ago: implications for global-scale plate tectonics Hamed Gamal El Dien1,2 ✉ , Luc S. Doucet1, J. Brendan Murphy1,3 & Zheng-Xiang Li1 Progressive mantle melting during the Earth’s earliest evolution led to the formation of a depleted mantle and a continental crust enriched in highly incompatible elements. Re-enrichment of Earth’s mantle can occur when continental crustal materials begin to founder into the mantle by either subduction or, to a lesser degree, by delamination processes, profoundly afecting the mantle’s trace element and volatile compositions. Deciphering when mantle re-enrichment/refertilization became a global-scale process would reveal the onset of efcient mass transfer of crust to the mantle and potentially when plate tectonic processes became operative on a global-scale. Here we document the onset of mantle re-enrichment/refertilization by comparing the abundances of petrogenetically signifcant isotopic values and key ratios of highly incompatible elements compared to lithophile elements in Archean to Early-Proterozoic mantle-derived melts (i.e., basalts and komatiites). Basalts and komatiites both record a rapid-change in mantle chemistry around 3.2 billion years ago (Ga) signifying a fundamental change in Earth geodynamics. This rapid-change is recorded in Nd isotopes and in key trace element ratios that refect a fundamental shift in the balance between fuid-mobile and incompatible elements (i.e., Ba/La, Ba/Nb, U/Nb, Pb/Nd and Pb/Ce) in basaltic and komatiitic rocks. These geochemical proxies display a signifcant increase in magnitude and variability after ~3.2 Ga. -

Appendix 1*) for Essential Information Very Well Illustrated in Google and Wikipedia
Appendix 1*) For Essential Information Very Well Illustrated in Google and Wikipedia *) in Support of the Text with Literature Citations. Referrals to illustrations in Appendix 2. Cancer in the Plant. The insertion of the Agrobacterium tumefaciens circular plasmid T (transferred) DNA into the genome of its new host, the plant (Gelvin BS. Microbiol Molecular Biol Rev 2003;67:16–37). The plant cancer “crown gall” (agrocallus; Agrobacterial crown gall) consists of malignantly transformed cells replicating the agrobacterial T DNA plasmid (reviewed in postscript Table XXXV). For reference: Koncz C Mayerhofer R Koncz-Kálmán Zs et al EMBO J 1990;9:1337–1346. Transfer of potentially oncogenic bacterial genes and proteins to patients: Septicemic Bacteroides enterotoxigenic (Sinkovics J G & Smith JP Cancer 1970;25:663–671; Viljoen KS et al PLoS One 2015;10(3):e0119462); Bartonella bacilliformis etc (Guy L et al PLoS Genet 2013;9(3):e1003393; Harms A & Dehio C Clin Microbiol Rev 2012;25:42–78; Llosa M et al Trends Microbiol 2012;20:355–9; Minnick MF et al PLoS Negl Trop Dis 2014;6(7):e2919); Helicobacter pylori (Bonsor DA et al J Biol Chem 2015;pii:jbc.M115.641829; Su YL et al J Immunol 2015;194:3997–4007; Vaziri F et al Pathog Dis 2015;73(3). pii.ftu021); Porphyromonas gingivalis (Katz J et al Int J Oral Sci 2011;3:209–215); Tuberculous infections with A. tumefaciens in patients (Ramirez FC et al Clin Infect Dis 1992;15:938–940). DNA-binding Antibodies. DNA- (or RNA-) binding proteins use zink finger motifs, leucine zippers and winged (beta-sheet loops) helix-turn helix motifs (HTH, two helices separated by the loop, RNA/DNA-binding domains) in recognition of RNA/DNA receptors for attachment. -

Petrogenesis of 3.15 Ga Old Banasandra Komatiites from the Dharwar Craton, India: Implications for Early Mantle Heterogeneity
Accepted Manuscript Petrogenesis of 3.15 Ga old Banasandra komatiites from the Dharwar craton, India: Implications for early mantle heterogeneity J.M. Maya, Rajneesh Bhutani, S. Balakrishnan, S. Rajee Sandhya PII: S1674-9871(16)30025-1 DOI: 10.1016/j.gsf.2016.03.007 Reference: GSF 443 To appear in: Geoscience Frontiers Received Date: 5 November 2015 Revised Date: 1 March 2016 Accepted Date: 3 March 2016 Please cite this article as: Maya, J.M., Bhutani, R., Balakrishnan, S., Sandhya, S.R., Petrogenesis of 3.15 Ga old Banasandra komatiites from the Dharwar craton, India: Implications for early mantle heterogeneity, Geoscience Frontiers (2016), doi: 10.1016/j.gsf.2016.03.007. This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. ACCEPTED MANUSCRIPT MANUSCRIPT ACCEPTED ACCEPTED MANUSCRIPT 1 Petrogenesis of 3.15 Ga old Banasandra komatiites from the Dharwar 2 craton, India: Implications for early mantle heterogeneity * 3 J. M. Maya, Rajneesh Bhutani , S. Balakrishnan, S. Rajee Sandhya 4 Department of Earth Sciences, Pondicherry University, Puducherry-605014, India 5 6 *Corresponding author. Phone: +919443636422; 7 E-mail address: [email protected] 8 9 MANUSCRIPT ACCEPTED ACCEPTED MANUSCRIPT 10 Abstract 11 Spinifex-textured, magnesian (MgO >25 wt.%) komatiites from Mesoarchean 12 Banasandra greenstone belt of the Sargur Group in the Dharwar craton, India were 13 analysed for major and trace elements and 147,146 Sm-143,142 Nd systematics to constrain 14 age, petrogenesis and to understand the evolution of Archean mantle. -

M.Sc. Microbiology (2019 ONWARDS)
PONDICHERRY UNIVERSITY PUDUCHERRY 605 014 CURRICULUM AND SYLLABUS of M.Sc. Microbiology (2019 ONWARDS) Department of Microbiology School of Life Sciences About the course The Department of Microbiology is committed to excellence in education, research and extension. This Department is being strengthened with various research units and periodical update / modernization of the curricula. The Department of Microbiology at the Pondicherry University, School of Life Sciences, brings together a variety of researchers as faculty of this programme who are specialized in their domains and united by the common goal of understanding the “Microbes”. Microbes are playing important role in the bioprocess of all living things and maintain homeostasis of the universe. Without microbes, one cannot imagine such a biologically balanced and diverse universe; rather our earth would have placed as a barren planet. As the microbial activities are so diverse, the microbiology programme is a multidisciplinary subject, which will have the roots of life science, environmental science, and engineering. Traditional microbiology is considered to be an important area of study in biology since it has enormous potential and vast scope in fermentation, bioremediation and biomedical technology. But the recent developments from human microbiome project, metagenomics and microbial genome projects has expanded its scope and potential in the next generation drug design, molecular pathogenesis, phylogeography, production of smart biomolecules, etc. Modern Microbiology has expanded its roots in genome technology, nanobiotechnology, green energy (biofuel) technology, bioelectronics etc. Considering recent innovations and rapid growth of microbiological approaches and applications in human and environmental sustainability, the M.Sc. Microbiology curricula is designed to enlighten the students in basics of Microbiology to recent developments. -

Geology of the Nuvvuagittuq Belt
Earth’s Oldest Rocks Edited by Martin J. van Kranendonk, R. Hugh Smithies and Vickie C. Bennett Developments in Precambrian Geology, Vol. 15 (K.C. Condie, Series Editor) ©2007 Elsevier B.V. All rights reserved. THE GEOLOGY OF THE 3.8 GA NUVVUAGITTUQ (PORPOISE COVE) GREENSTONE BELT, NORTHEASTERN SUPERIOR PROVINCE, CANADA. Jonathan O’Neil1, Charles Maurice2, Ross K. Stevenson3,4, Jeff Larocque5, Christophe Cloquet6, Jean David3 & Don Francis1 1 Earth & Planetary Sciences, McGill University and GÉOTOP-UQÀM-McGill, 3450 University St. Montreal, QC, Canada, H3A 2A7 ([email protected]) 2 Bureau de l’exploration géologique du Québec, Ministère des Ressources naturelles et de la Faune, 400 boul. Lamaque Val d’Or, QC, J9P 3L4 3 GÉOTOP-UQÀM-McGill, Université du Québec à Montréal, C.P. 8888, succ. centre-ville, Montreal, QC, Canada H3C 3P8 4 Département des Sciences de la Terre et de l’Atmosphère, Université du Québec à Montréal, C.P. 8888, succ. centre-ville Montreal, QC, Canada H3C 3P8 5 School of Earth and Oceanic Sciences, University of Victoria, P.O. Box 3055 STN CSC, Victoria, BC, Canada, V8W 3P6 6 INW-UGent, Department of analytical chemistry, Proeftuinstraat 86, 9000 GENT, Belgium Abstract The Nuvvuagittuq greenstone belt is a 3.8 Ga supracrustal succession preserved as a raft in remobilised tonalities along the eastern coast of Hudson Bay. The dominant lithology of the belt is a quartz-ribboned grey amphibolite composed of variable proportions of cummingtonite, biotite and plagioclase. Although the amphibolites have mafic compositions, the presence of cummingtonite rather than hornblende in these rocks reflects their low Ca contents, which may result from the alteration and metamorphism of mafic pyroclastic rocks. -

RESEARCH a Non–Plate Tectonic
RESEARCH A non–plate tectonic model for the Eoarchean Isua supracrustal belt A. Alexander G. Webb1,*, Thomas Müller2, Jiawei Zuo1, Peter J. Haproff3, and Anthony Ramírez-Salazar2 1DEPARTMENT OF EARTH SCIENCES AND LABORATORY FOR SPACE RESEARCH, UNIVERSITY OF HONG KONG, POKFULAM ROAD, HONG KONG, CHINA 2SCHOOL OF EARTH AND ENVIRONMENT, UNIVERSITY OF LEEDS, MATHS/EARTH AND ENVIRONMENT BUILDING, LEEDS LS2 9JT, UK 3DEPARTMENT OF EARTH AND OCEAN SCIENCES, UNIVERSITY OF NORTH CAROLINA, WILMINGTON, NORTH CAROLINA 28403, USA ABSTRACT The ca. 3.8–3.6-b.y.-old Isua supracrustal belt of SW Greenland is Earth’s only site older than 3.2 Ga that is exclusively interpreted via plate- tectonic theory. The belt is divided into ca. 3.8 Ga and ca. 3.7 Ga halves, and these are interpreted as plate fragments that collided by ca. 3.6 Ga. However, such models are based on idiosyncratic interpretations of field observations and U-Pb zircon data, resulting in intricate, conflicting stratigraphic and structural interpretations. We reanalyzed published geochronological work and associated field constraints previously interpreted to show multiple plate-tectonic events and conducted field-based exploration of metamorphic and structural gra- dients previously interpreted to show heterogeneities recording plate-tectonic processes. Simpler interpretations are viable, i.e., the belt may have experienced nearly homogeneous metamorphic conditions and strain during a single deformation event prior to intrusion of ca. 3.5 Ga mafic dikes. Curtain and sheath folds occur at multiple scales throughout the belt, with the entire belt potentially representing Earth’s largest a-type fold. Integrating these findings, we present a new model in which two cycles of volcanic burial and resultant melt- ing and tonalite-trondhjemite-granodiorite (TTG) intrusion produced first the ca.