Functional in Vivo Screen Identifies Pygo2 As a Putative Gene to Promote Prostate Cancer
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Constitutive Scaffolding of Multiple Wnt Enhanceosome Components By
RESEARCH ARTICLE Constitutive scaffolding of multiple Wnt enhanceosome components by Legless/ BCL9 Laurens M van Tienen, Juliusz Mieszczanek, Marc Fiedler, Trevor J Rutherford, Mariann Bienz* MRC Laboratory of Molecular Biology, Cambridge, United Kingdom Abstract Wnt/b-catenin signaling elicits context-dependent transcription switches that determine normal development and oncogenesis. These are mediated by the Wnt enhanceosome, a multiprotein complex binding to the Pygo chromatin reader and acting through TCF/LEF- responsive enhancers. Pygo renders this complex Wnt-responsive, by capturing b-catenin via the Legless/BCL9 adaptor. We used CRISPR/Cas9 genome engineering of Drosophila legless (lgs) and human BCL9 and B9L to show that the C-terminus downstream of their adaptor elements is crucial for Wnt responses. BioID proximity labeling revealed that BCL9 and B9L, like PYGO2, are constitutive components of the Wnt enhanceosome. Wnt-dependent docking of b-catenin to the enhanceosome apparently causes a rearrangement that apposes the BCL9/B9L C-terminus to TCF. This C-terminus binds to the Groucho/TLE co-repressor, and also to the Chip/LDB1-SSDP enhanceosome core complex via an evolutionary conserved element. An unexpected link between BCL9/B9L, PYGO2 and nuclear co-receptor complexes suggests that these b-catenin co-factors may coordinate Wnt and nuclear hormone responses. DOI: 10.7554/eLife.20882.001 *For correspondence: mb2@mrc- Introduction lmb.cam.ac.uk The Wnt/b-catenin signaling cascade is an ancient cell communication pathway that operates con- Competing interests: The text-dependent transcriptional switches to control animal development and tissue homeostasis authors declare that no (Cadigan and Nusse, 1997). -

A Gene-Level Methylome-Wide Association Analysis Identifies Novel
bioRxiv preprint doi: https://doi.org/10.1101/2020.07.13.201376; this version posted July 14, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 A gene-level methylome-wide association analysis identifies novel 2 Alzheimer’s disease genes 1 1 2 3 4 3 Chong Wu , Jonathan Bradley , Yanming Li , Lang Wu , and Hong-Wen Deng 1 4 Department of Statistics, Florida State University; 2 5 Department of Biostatistics & Data Science, University of Kansas Medical Center; 3 6 Population Sciences in the Pacific Program, University of Hawaii Cancer center; 4 7 Tulane Center for Biomedical Informatics and Genomics, Deming Department of Medicine, 8 Tulane University School of Medicine 9 Corresponding to: Chong Wu, Assistant Professor, Department of Statistics, Florida State 10 University, email: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.07.13.201376; this version posted July 14, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 11 Abstract 12 Motivation: Transcriptome-wide association studies (TWAS) have successfully facilitated the dis- 13 covery of novel genetic risk loci for many complex traits, including late-onset Alzheimer’s disease 14 (AD). However, most existing TWAS methods rely only on gene expression and ignore epige- 15 netic modification (i.e., DNA methylation) and functional regulatory information (i.e., enhancer- 16 promoter interactions), both of which contribute significantly to the genetic basis ofAD. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Dynamic Transcriptomic Profiles of Zebrafish Gills in Response to Zinc
Zheng et al. BMC Genomics 2010, 11:548 http://www.biomedcentral.com/1471-2164/11/548 RESEARCH ARTICLE Open Access Dynamic transcriptomic profiles of zebrafish gills in response to zinc depletion Dongling Zheng1,4, Peter Kille2, Graham P Feeney2, Phil Cunningham1, Richard D Handy3, Christer Hogstrand1* Abstract Background: Zinc deficiency is detrimental to organisms, highlighting its role as an essential micronutrient contributing to numerous biological processes. To investigate the underlying molecular events invoked by zinc depletion we performed a temporal analysis of transcriptome changes observed within the zebrafish gill. This tissue represents a model system for studying ion absorption across polarised epithelial cells as it provides a major pathway for fish to acquire zinc directly from water whilst sharing a conserved zinc transporting system with mammals. Results: Zebrafish were treated with either zinc-depleted (water = 2.61 μgL-1; diet = 26 mg kg-1) or zinc-adequate (water = 16.3 μgL-1; diet = 233 mg kg-1) conditions for two weeks. Gill samples were collected at five time points and transcriptome changes analysed in quintuplicate using a 16K oligonucleotide array. Of the genes represented the expression of a total of 333 transcripts showed differential regulation by zinc depletion (having a fold-change greater than 1.8 and an adjusted P-value less than 0.1, controlling for a 10% False Discovery Rate). Down-regulation was dominant at most time points and distinct sets of genes were regulated at different stages. Annotation enrichment analysis revealed that ‘Developmental Process’ was the most significantly overrepresented Biological Process GO term (P = 0.0006), involving 26% of all regulated genes. -

The Interactome of KRAB Zinc Finger Proteins Reveals the Evolutionary History of Their Functional Diversification
Resource The interactome of KRAB zinc finger proteins reveals the evolutionary history of their functional diversification Pierre-Yves Helleboid1,†, Moritz Heusel2,†, Julien Duc1, Cécile Piot1, Christian W Thorball1, Andrea Coluccio1, Julien Pontis1, Michaël Imbeault1, Priscilla Turelli1, Ruedi Aebersold2,3,* & Didier Trono1,** Abstract years ago (MYA) (Imbeault et al, 2017). Their products harbor an N-terminal KRAB (Kru¨ppel-associated box) domain related to that of Krüppel-associated box (KRAB)-containing zinc finger proteins Meisetz (a.k.a. PRDM9), a protein that originated prior to the diver- (KZFPs) are encoded in the hundreds by the genomes of higher gence of chordates and echinoderms, and a C-terminal array of zinc vertebrates, and many act with the heterochromatin-inducing fingers (ZNF) with sequence-specific DNA-binding potential (Urru- KAP1 as repressors of transposable elements (TEs) during early tia, 2003; Birtle & Ponting, 2006; Imbeault et al, 2017). KZFP genes embryogenesis. Yet, their widespread expression in adult tissues multiplied by gene and segment duplication to count today more and enrichment at other genetic loci indicate additional roles. than 350 and 700 representatives in the human and mouse Here, we characterized the protein interactome of 101 of the ~350 genomes, respectively (Urrutia, 2003; Kauzlaric et al, 2017). A human KZFPs. Consistent with their targeting of TEs, most KZFPs majority of human KZFPs including all primate-restricted family conserved up to placental mammals essentially recruit KAP1 and members target sequences derived from TEs, that is, DNA trans- associated effectors. In contrast, a subset of more ancient KZFPs posons, ERVs (endogenous retroviruses), LINEs, SINEs (long and rather interacts with factors related to functions such as genome short interspersed nuclear elements, respectively), or SVAs (SINE- architecture or RNA processing. -

Table S1| Differential Expression Analysis of the Atopy Transcriptome
Table S1| Differential expression analysis of the atopy transcriptome in CD4+ T-cell responses to allergens in atopic and nonatopic subjects Probe ID S.test Gene Symbol Gene Description Chromosome Statistic Location 7994280 10.32 IL4R Interleukin 4 receptor 16p11.2-12.1 8143383 8.95 --- --- --- 7974689 8.50 DACT1 Dapper, antagonist of beta-catenin, homolog 1 14q23.1 8102415 7.59 CAMK2D Calcium/calmodulin-dependent protein kinase II delta 4q26 7950743 7.58 RAB30 RAB30, member RAS oncogene family 11q12-q14 8136580 7.54 RAB19B GTP-binding protein RAB19B 7q34 8043504 7.45 MAL Mal, T-cell differentiation protein 2cen-q13 8087739 7.27 CISH Cytokine inducible SH2-containing protein 3p21.3 8000413 7.17 NSMCE1 Non-SMC element 1 homolog (S. cerevisiae) 16p12.1 8021301 7.15 RAB27B RAB27B, member RAS oncogene family 18q21.2 8143367 6.83 SLC37A3 Solute carrier family 37 member 3 7q34 8152976 6.65 TMEM71 Transmembrane protein 71 8q24.22 7931914 6.56 IL2R Interleukin 2 receptor, alpha 10p15-p14 8014768 6.43 PLXDC1 Plexin domain containing 1 17q21.1 8056222 6.43 DPP4 Dipeptidyl-peptidase 4 (CD26) 2q24.3 7917697 6.40 GFI1 Growth factor independent 1 1p22 7903507 6.39 FAM102B Family with sequence similarity 102, member B 1p13.3 7968236 5.96 RASL11A RAS-like, family 11, member A --- 7912537 5.95 DHRS3 Dehydrogenase/reductase (SDR family) member 3 1p36.1 7963491 5.83 KRT1 Keratin 1 (epidermolytic hyperkeratosis) 12q12-q13 7903786 5.72 CSF1 Colony stimulating factor 1 (macrophage) 1p21-p13 8019061 5.67 SGSH N-sulfoglucosamine sulfohydrolase (sulfamidase) 17q25.3 -

Inhibition of Nuclear Wnt Signaling: Challenges of an Elusive Target for Cancer Therapy
1 Inhibition of Nuclear Wnt Signaling: Challenges of an Elusive Target for Cancer Therapy Yung Lyou1*, Amber N. Habowski2, George T. Chen2, Marian L. Waterman2*. 1University of California Irvine Medical Center, Department of Medicine, Division of Hematology Oncology, 101 The City Drive South, Orange, CA, USA, 92868; 2University of California Irvine, Department of Microbiology and Molecular Genetics, Irvine, CA, USA, 92697. *Co-corresponding authors 2 Abstract The highly conserved Wnt signaling pathway plays an important role in embryonic development and disease pathogenesis, most notably cancer. The “canonical,” or - catenin-dependent Wnt signal initiates at the cell plasma membrane with the binding of Wnt proteins to Frizzled:LRP5/LRP6 receptor complexes, and is mediated by the translocation of the transcription co-activator protein, -catenin, into the nucleus. -catenin then forms a complex with TCF/LEF transcription factors to regulate multiple gene programs. These programs play roles in cell proliferation, migration, vasculogenesis, survival, and metabolism. Mutations in Wnt signaling pathway components that lead to constitutively active Wnt signaling drive aberrant expression of these programs and development of cancer. It has been a longstanding and challenging goal to develop therapies that can interfere with the TCF/LEF--catenin transcriptional complex. This review will focus on the i) structural considerations for targeting the TCF/LEF--catenin and co-regulatory complexes in the nucleus, ii) current molecules that directly target -

X-Linked Diseases: Susceptible Females
REVIEW ARTICLE X-linked diseases: susceptible females Barbara R. Migeon, MD 1 The role of X-inactivation is often ignored as a prime cause of sex data include reasons why women are often protected from the differences in disease. Yet, the way males and females express their deleterious variants carried on their X chromosome, and the factors X-linked genes has a major role in the dissimilar phenotypes that that render women susceptible in some instances. underlie many rare and common disorders, such as intellectual deficiency, epilepsy, congenital abnormalities, and diseases of the Genetics in Medicine (2020) 22:1156–1174; https://doi.org/10.1038/s41436- heart, blood, skin, muscle, and bones. Summarized here are many 020-0779-4 examples of the different presentations in males and females. Other INTRODUCTION SEX DIFFERENCES ARE DUE TO X-INACTIVATION Sex differences in human disease are usually attributed to The sex differences in the effect of X-linked pathologic variants sex specific life experiences, and sex hormones that is due to our method of X chromosome dosage compensation, influence the function of susceptible genes throughout the called X-inactivation;9 humans and most placental mammals – genome.1 5 Such factors do account for some dissimilarities. compensate for the sex difference in number of X chromosomes However, a major cause of sex-determined expression of (that is, XX females versus XY males) by transcribing only one disease has to do with differences in how males and females of the two female X chromosomes. X-inactivation silences all X transcribe their gene-rich human X chromosomes, which is chromosomes but one; therefore, both males and females have a often underappreciated as a cause of sex differences in single active X.10,11 disease.6 Males are the usual ones affected by X-linked For 46 XY males, that X is the only one they have; it always pathogenic variants.6 Females are biologically superior; a comes from their mother, as fathers contribute their Y female usually has no disease, or much less severe disease chromosome. -

Elucidating the Roles of Wnt-Secretion and Β-Catenin's
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2018 Elucidating the roles of Wnt-secretion and -catenin’s interaction partners in development and disease Zimmerli, Dario Posted at the Zurich Open Repository and Archive, University of Zurich ZORA URL: https://doi.org/10.5167/uzh-165192 Dissertation Published Version Originally published at: Zimmerli, Dario. Elucidating the roles of Wnt-secretion and -catenin’s interaction partners in develop- ment and disease. 2018, University of Zurich, Faculty of Science. Elucidating the Roles of Wnt-Secretion and b-Catenin’s Interaction Partners in Development and Disease Dissertation zur Erlangung der naturwissenschaftlichen Doktorwürde (Dr. sc. nat.) vorgelegt der Mathematisch-naturwissenschaftlichen Fakultät der Universität Zürich von Dario Zimmerli von Oftringen, Aargau Promotionskommission Prof. Dr. Konrad Basler (Vorsitz und Leitung der Dissertation) Prof. Dr. Maries van den BroeK Prof. Dr. Sabine Werner Zürich, 2018 Summary ........................................................................................................................... 3 Zusammenfassung ............................................................................................................ 5 Introduction ...................................................................................................................... 7 Review ........................................................................................................................... -
Download Validation Data
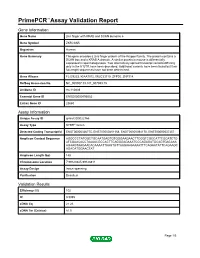
PrimePCR™Assay Validation Report Gene Information Gene Name zinc finger with KRAB and SCAN domains 5 Gene Symbol ZKSCAN5 Organism Human Gene Summary This gene encodes a zinc finger protein of the Kruppel family. The protein contains a SCAN box and a KRAB A domain. A similar protein in mouse is differentially expressed in spermatogenesis. Two alternatively spliced transcript variants differing only in the 5' UTR have been described. Additional variants have been found but their full-length sequences have not been determined. Gene Aliases FLJ39233, KIAA1015, MGC33710, ZFP95, ZNF914 RefSeq Accession No. NC_000007.13, NT_007933.15 UniGene ID Hs.110839 Ensembl Gene ID ENSG00000196652 Entrez Gene ID 23660 Assay Information Unique Assay ID qHsaCID0022766 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000326775, ENST00000451158, ENST00000394170, ENST00000537357 Amplicon Context Sequence AGGCCCTATGGCTGCAATGAGTGTGGGAAGAACTTCGGTCGCCATTCGCATCTG ATCGAACACCTAAAACGCCACTTCAGGGAGAAATCCCAGAGATGCAGTGACAAA AGAAGTAAGAACACAAAATTAAGTGTTAAGAAGAAAATTTCAGAATATTCAGAAGC AGACATGGAACTAT Amplicon Length (bp) 148 Chromosome Location 7:99123945-99128811 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 102 R2 0.9985 cDNA Cq 21.26 cDNA Tm (Celsius) 81.5 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq 34.61 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report ZKSCAN5, Human Amplification Plot Amplification of cDNA generated from -

Supplementary Table 2
Supplementary Table 2. Differentially Expressed Genes following Sham treatment relative to Untreated Controls Fold Change Accession Name Symbol 3 h 12 h NM_013121 CD28 antigen Cd28 12.82 BG665360 FMS-like tyrosine kinase 1 Flt1 9.63 NM_012701 Adrenergic receptor, beta 1 Adrb1 8.24 0.46 U20796 Nuclear receptor subfamily 1, group D, member 2 Nr1d2 7.22 NM_017116 Calpain 2 Capn2 6.41 BE097282 Guanine nucleotide binding protein, alpha 12 Gna12 6.21 NM_053328 Basic helix-loop-helix domain containing, class B2 Bhlhb2 5.79 NM_053831 Guanylate cyclase 2f Gucy2f 5.71 AW251703 Tumor necrosis factor receptor superfamily, member 12a Tnfrsf12a 5.57 NM_021691 Twist homolog 2 (Drosophila) Twist2 5.42 NM_133550 Fc receptor, IgE, low affinity II, alpha polypeptide Fcer2a 4.93 NM_031120 Signal sequence receptor, gamma Ssr3 4.84 NM_053544 Secreted frizzled-related protein 4 Sfrp4 4.73 NM_053910 Pleckstrin homology, Sec7 and coiled/coil domains 1 Pscd1 4.69 BE113233 Suppressor of cytokine signaling 2 Socs2 4.68 NM_053949 Potassium voltage-gated channel, subfamily H (eag- Kcnh2 4.60 related), member 2 NM_017305 Glutamate cysteine ligase, modifier subunit Gclm 4.59 NM_017309 Protein phospatase 3, regulatory subunit B, alpha Ppp3r1 4.54 isoform,type 1 NM_012765 5-hydroxytryptamine (serotonin) receptor 2C Htr2c 4.46 NM_017218 V-erb-b2 erythroblastic leukemia viral oncogene homolog Erbb3 4.42 3 (avian) AW918369 Zinc finger protein 191 Zfp191 4.38 NM_031034 Guanine nucleotide binding protein, alpha 12 Gna12 4.38 NM_017020 Interleukin 6 receptor Il6r 4.37 AJ002942 -

Exons Deletion of CNKSR2 Gene Identified in X-Linked Syndromic
Daoqi et al. BMC Medical Genetics (2020) 21:69 https://doi.org/10.1186/s12881-020-01004-2 CASE REPORT Open Access Exons deletion of CNKSR2 gene identified in X-linked syndromic intellectual disability Mei Daoqi1, Chen Guohong1, Wang Yuan1, Yang Zhixiao1, Xu Kaili1 and Mei Shiyue2* Abstract Background: The Houge type of X-linked syndromic mental retardation is an X-linked intellectual disability (XLID) recently recorded in the Online Mendelian Inheritance in Man (OMIM) and only 8 cases have been reported in literature thus far. Case presentation: We present two brothers with intractable seizures and syndromic intellectual disability with symptoms consisting of delayed development, intellectual disability, and speech and language delay. The mother was a symptomatic carrier with milder clinical phenotype. Whole exome sequencing identified a small fragment deletion spanning four exons, about 9.5 kilobases (kb) in length in the CNKSR2 gene in the patients. The mutation co-segregation revealed that exon deletions occurred de novo in the proband’s mother. Conclusion: Although large deletions have been reported, no small deletions have yet been identified. In this case report, we identified a small deletion in the CNKSR2 gene. This study enhances our knowledge of the CNKSR2 gene mutation spectrum and provides further information about the phenotypic characteristics of X-linked syndromic intellectual disability. Keywords: X-linked syndromic mental retardation, Intractable seizures, CNKSR2 gene, Exons deletion Background (#301008) in 2017. MRXSHG is caused by a deficiency X-linked intellectual disability (XLID), also known as of the CNKSR2 gene and only 8 cases have been X-linked mental retardation (XLMR), refers to a gener- reported in literature.