Eight Common Genetic Variants Associated with Serum DHEAS Levels Suggest a Key Role in Ageing Mechanisms
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Dual Proteome-Scale Networks Reveal Cell-Specific Remodeling of the Human Interactome
bioRxiv preprint doi: https://doi.org/10.1101/2020.01.19.905109; this version posted January 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Dual Proteome-scale Networks Reveal Cell-specific Remodeling of the Human Interactome Edward L. Huttlin1*, Raphael J. Bruckner1,3, Jose Navarrete-Perea1, Joe R. Cannon1,4, Kurt Baltier1,5, Fana Gebreab1, Melanie P. Gygi1, Alexandra Thornock1, Gabriela Zarraga1,6, Stanley Tam1,7, John Szpyt1, Alexandra Panov1, Hannah Parzen1,8, Sipei Fu1, Arvene Golbazi1, Eila Maenpaa1, Keegan Stricker1, Sanjukta Guha Thakurta1, Ramin Rad1, Joshua Pan2, David P. Nusinow1, Joao A. Paulo1, Devin K. Schweppe1, Laura Pontano Vaites1, J. Wade Harper1*, Steven P. Gygi1*# 1Department of Cell Biology, Harvard Medical School, Boston, MA, 02115, USA. 2Broad Institute, Cambridge, MA, 02142, USA. 3Present address: ICCB-Longwood Screening Facility, Harvard Medical School, Boston, MA, 02115, USA. 4Present address: Merck, West Point, PA, 19486, USA. 5Present address: IQ Proteomics, Cambridge, MA, 02139, USA. 6Present address: Vor Biopharma, Cambridge, MA, 02142, USA. 7Present address: Rubius Therapeutics, Cambridge, MA, 02139, USA. 8Present address: RPS North America, South Kingstown, RI, 02879, USA. *Correspondence: [email protected] (E.L.H.), [email protected] (J.W.H.), [email protected] (S.P.G.) #Lead Contact: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2020.01.19.905109; this version posted January 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. -

A Gene-Level Methylome-Wide Association Analysis Identifies Novel
bioRxiv preprint doi: https://doi.org/10.1101/2020.07.13.201376; this version posted July 14, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 A gene-level methylome-wide association analysis identifies novel 2 Alzheimer’s disease genes 1 1 2 3 4 3 Chong Wu , Jonathan Bradley , Yanming Li , Lang Wu , and Hong-Wen Deng 1 4 Department of Statistics, Florida State University; 2 5 Department of Biostatistics & Data Science, University of Kansas Medical Center; 3 6 Population Sciences in the Pacific Program, University of Hawaii Cancer center; 4 7 Tulane Center for Biomedical Informatics and Genomics, Deming Department of Medicine, 8 Tulane University School of Medicine 9 Corresponding to: Chong Wu, Assistant Professor, Department of Statistics, Florida State 10 University, email: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.07.13.201376; this version posted July 14, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 11 Abstract 12 Motivation: Transcriptome-wide association studies (TWAS) have successfully facilitated the dis- 13 covery of novel genetic risk loci for many complex traits, including late-onset Alzheimer’s disease 14 (AD). However, most existing TWAS methods rely only on gene expression and ignore epige- 15 netic modification (i.e., DNA methylation) and functional regulatory information (i.e., enhancer- 16 promoter interactions), both of which contribute significantly to the genetic basis ofAD. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Dynamic Transcriptomic Profiles of Zebrafish Gills in Response to Zinc
Zheng et al. BMC Genomics 2010, 11:548 http://www.biomedcentral.com/1471-2164/11/548 RESEARCH ARTICLE Open Access Dynamic transcriptomic profiles of zebrafish gills in response to zinc depletion Dongling Zheng1,4, Peter Kille2, Graham P Feeney2, Phil Cunningham1, Richard D Handy3, Christer Hogstrand1* Abstract Background: Zinc deficiency is detrimental to organisms, highlighting its role as an essential micronutrient contributing to numerous biological processes. To investigate the underlying molecular events invoked by zinc depletion we performed a temporal analysis of transcriptome changes observed within the zebrafish gill. This tissue represents a model system for studying ion absorption across polarised epithelial cells as it provides a major pathway for fish to acquire zinc directly from water whilst sharing a conserved zinc transporting system with mammals. Results: Zebrafish were treated with either zinc-depleted (water = 2.61 μgL-1; diet = 26 mg kg-1) or zinc-adequate (water = 16.3 μgL-1; diet = 233 mg kg-1) conditions for two weeks. Gill samples were collected at five time points and transcriptome changes analysed in quintuplicate using a 16K oligonucleotide array. Of the genes represented the expression of a total of 333 transcripts showed differential regulation by zinc depletion (having a fold-change greater than 1.8 and an adjusted P-value less than 0.1, controlling for a 10% False Discovery Rate). Down-regulation was dominant at most time points and distinct sets of genes were regulated at different stages. Annotation enrichment analysis revealed that ‘Developmental Process’ was the most significantly overrepresented Biological Process GO term (P = 0.0006), involving 26% of all regulated genes. -

The Interactome of KRAB Zinc Finger Proteins Reveals the Evolutionary History of Their Functional Diversification
Resource The interactome of KRAB zinc finger proteins reveals the evolutionary history of their functional diversification Pierre-Yves Helleboid1,†, Moritz Heusel2,†, Julien Duc1, Cécile Piot1, Christian W Thorball1, Andrea Coluccio1, Julien Pontis1, Michaël Imbeault1, Priscilla Turelli1, Ruedi Aebersold2,3,* & Didier Trono1,** Abstract years ago (MYA) (Imbeault et al, 2017). Their products harbor an N-terminal KRAB (Kru¨ppel-associated box) domain related to that of Krüppel-associated box (KRAB)-containing zinc finger proteins Meisetz (a.k.a. PRDM9), a protein that originated prior to the diver- (KZFPs) are encoded in the hundreds by the genomes of higher gence of chordates and echinoderms, and a C-terminal array of zinc vertebrates, and many act with the heterochromatin-inducing fingers (ZNF) with sequence-specific DNA-binding potential (Urru- KAP1 as repressors of transposable elements (TEs) during early tia, 2003; Birtle & Ponting, 2006; Imbeault et al, 2017). KZFP genes embryogenesis. Yet, their widespread expression in adult tissues multiplied by gene and segment duplication to count today more and enrichment at other genetic loci indicate additional roles. than 350 and 700 representatives in the human and mouse Here, we characterized the protein interactome of 101 of the ~350 genomes, respectively (Urrutia, 2003; Kauzlaric et al, 2017). A human KZFPs. Consistent with their targeting of TEs, most KZFPs majority of human KZFPs including all primate-restricted family conserved up to placental mammals essentially recruit KAP1 and members target sequences derived from TEs, that is, DNA trans- associated effectors. In contrast, a subset of more ancient KZFPs posons, ERVs (endogenous retroviruses), LINEs, SINEs (long and rather interacts with factors related to functions such as genome short interspersed nuclear elements, respectively), or SVAs (SINE- architecture or RNA processing. -

WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT (51) International Patent Classification: CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, C12Q 1/68 (2018.01) A61P 31/18 (2006.01) DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, C12Q 1/70 (2006.01) HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, (21) International Application Number: MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, PCT/US2018/056167 OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, (22) International Filing Date: SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, 16 October 2018 (16. 10.2018) TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (26) Publication Language: English GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, TZ, (30) Priority Data: UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, 62/573,025 16 October 2017 (16. 10.2017) US TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, EE, ES, FI, FR, GB, GR, HR, HU, ΓΕ , IS, IT, LT, LU, LV, (71) Applicant: MASSACHUSETTS INSTITUTE OF MC, MK, MT, NL, NO, PL, PT, RO, RS, SE, SI, SK, SM, TECHNOLOGY [US/US]; 77 Massachusetts Avenue, TR), OAPI (BF, BJ, CF, CG, CI, CM, GA, GN, GQ, GW, Cambridge, Massachusetts 02139 (US). -

ARPC1A Antibody Cat
ARPC1A Antibody Cat. No.: 42-980 ARPC1A Antibody 42-980 (0.1ug/ml) staining of fetal Mouse Brain (A) and adult Rat Brain (B) lysates (35ug protein 42-980 (5ug/ml) staining of paraffin embedded Human Heart. in RIPA buffer). Primary incubation was 1 hour. Steamed antigen retrieval with citrate buffer pH 6, AP-staining. Detected by chemiluminescence. Specifications HOST SPECIES: Goat SPECIES REACTIVITY: Human, Mouse, Rat HOMOLOGY: Expected Species Reactivity based on sequence homology: Pig, Cow IMMUNOGEN: The immunogen for this antibody is: C-DFKTLESSIQGLR TESTED APPLICATIONS: ELISA, IHC, WB September 26, 2021 1 https://www.prosci-inc.com/arpc1a-antibody-42-980.html Peptide ELISA: antibody detection limit dilution 1:64000.Western Blot:Approx 45kDa band observed in lysates of cell line HeLa (calculated MW of 41.6kDa according to NP_006400.2). Approx 37kDa band observed in fetal Mouse Brain and adult Rat Brain APPLICATIONS: lysates (calculated MW of 41.6kDa according to Mouse NP_062741.1). Recommended concentration: 0.1-0.3ug/ml.Immunohistochemistry:Paraffin embedded Human Heart. Recommended concentration: 5ug/ml. This antibody is expected to recognize both reported isoforms (NP_006400.2; SPECIFICITY: NP_001177925.1). No cross-reactiviry expected with ARPC1B. PREDICTED MOLECULAR Approx 45 kDa WEIGHT: Properties Purified from goat serum by ammonium sulphate precipitation followed by antigen PURIFICATION: affinity chromatography using the immunizing peptide. CLONALITY: Polyclonal CONJUGATE: Unconjugated PHYSICAL STATE: Liquid Supplied at 0.5 mg/ml in Tris saline, 0.02% sodium azide, pH7.3 with 0.5% bovine serum BUFFER: albumin. Aliquot and store at -20°C. Minimize freezing and thawing. CONCENTRATION: 500 ug/mL STORAGE CONDITIONS: Aliquot and store at -20˚C. -

Integrating Protein Copy Numbers with Interaction Networks to Quantify Stoichiometry in Mammalian Endocytosis
bioRxiv preprint doi: https://doi.org/10.1101/2020.10.29.361196; this version posted October 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Integrating protein copy numbers with interaction networks to quantify stoichiometry in mammalian endocytosis Daisy Duan1, Meretta Hanson1, David O. Holland2, Margaret E Johnson1* 1TC Jenkins Department of Biophysics, Johns Hopkins University, 3400 N Charles St, Baltimore, MD 21218. 2NIH, Bethesda, MD, 20892. *Corresponding Author: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2020.10.29.361196; this version posted October 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Abstract Proteins that drive processes like clathrin-mediated endocytosis (CME) are expressed at various copy numbers within a cell, from hundreds (e.g. auxilin) to millions (e.g. clathrin). Between cell types with identical genomes, copy numbers further vary significantly both in absolute and relative abundance. These variations contain essential information about each protein’s function, but how significant are these variations and how can they be quantified to infer useful functional behavior? Here, we address this by quantifying the stoichiometry of proteins involved in the CME network. We find robust trends across three cell types in proteins that are sub- vs super-stoichiometric in terms of protein function, network topology (e.g. -
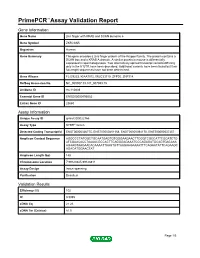
Download Validation Data
PrimePCR™Assay Validation Report Gene Information Gene Name zinc finger with KRAB and SCAN domains 5 Gene Symbol ZKSCAN5 Organism Human Gene Summary This gene encodes a zinc finger protein of the Kruppel family. The protein contains a SCAN box and a KRAB A domain. A similar protein in mouse is differentially expressed in spermatogenesis. Two alternatively spliced transcript variants differing only in the 5' UTR have been described. Additional variants have been found but their full-length sequences have not been determined. Gene Aliases FLJ39233, KIAA1015, MGC33710, ZFP95, ZNF914 RefSeq Accession No. NC_000007.13, NT_007933.15 UniGene ID Hs.110839 Ensembl Gene ID ENSG00000196652 Entrez Gene ID 23660 Assay Information Unique Assay ID qHsaCID0022766 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000326775, ENST00000451158, ENST00000394170, ENST00000537357 Amplicon Context Sequence AGGCCCTATGGCTGCAATGAGTGTGGGAAGAACTTCGGTCGCCATTCGCATCTG ATCGAACACCTAAAACGCCACTTCAGGGAGAAATCCCAGAGATGCAGTGACAAA AGAAGTAAGAACACAAAATTAAGTGTTAAGAAGAAAATTTCAGAATATTCAGAAGC AGACATGGAACTAT Amplicon Length (bp) 148 Chromosome Location 7:99123945-99128811 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 102 R2 0.9985 cDNA Cq 21.26 cDNA Tm (Celsius) 81.5 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq 34.61 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report ZKSCAN5, Human Amplification Plot Amplification of cDNA generated from -

ARPC1A Antibody (Center) Purified Rabbit Polyclonal Antibody (Pab) Catalog # Ap6519c
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 ARPC1A Antibody (Center) Purified Rabbit Polyclonal Antibody (Pab) Catalog # AP6519c Specification ARPC1A Antibody (Center) - Product Information Application WB, IHC-P, FC,E Primary Accession Q92747 Other Accession Q99PD4, Q9R0Q6, Q1JP79, Q8AVT9, A0A1L8EXB5 Reactivity Human Predicted Xenopus, Bovine, Mouse, Rat Host Rabbit Clonality Polyclonal Isotype Rabbit Ig Calculated MW 41569 Antigen Region 157-184 Western blot analysis of ARPC1A antibody ARPC1A Antibody (Center) - Additional (Center) (Cat.# AP6519c) in Y79 cell line Information lysates (35ug/lane). ARPC1A (arrow) was detected using the purified Pab. Gene ID 10552 Other Names Actin-related protein 2/3 complex subunit 1A, SOP2-like protein, ARPC1A, SOP2L Target/Specificity This ARPC1A antibody is generated from rabbits immunized with a KLH conjugated synthetic peptide between 157-184 amino acids from the Central region of human ARPC1A. Dilution WB~~1:1000 ARPC1A Antibody (Center) (Cat. #AP6519c) IHC-P~~1:50~100 immunohistochemistry analysis in formalin FC~~1:10~50 fixed and paraffin embedded human brain tissue followed by peroxidase conjugation of Format the secondary antibody and DAB staining. Purified polyclonal antibody supplied in PBS This data demonstrates the use of the with 0.09% (W/V) sodium azide. This ARPC1A Antibody (Center) for antibody is prepared by Saturated immunohistochemistry. Clinical relevance has Ammonium Sulfate (SAS) precipitation not been evaluated. followed by dialysis against PBS. Storage Maintain refrigerated at 2-8°C for up to 2 weeks. For long term storage store at -20°C Page 1/2 10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 in small aliquots to prevent freeze-thaw cycles. -

ARPC1A (NM 001190996) Human Tagged ORF Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC230814L4 ARPC1A (NM_001190996) Human Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: ARPC1A (NM_001190996) Human Tagged ORF Clone Tag: mGFP Symbol: ARPC1A Synonyms: Arc40; HEL-68; HEL-S-307; SOP2Hs; SOP2L Vector: pLenti-C-mGFP-P2A-Puro (PS100093) E. coli Selection: Chloramphenicol (34 ug/mL) Cell Selection: Puromycin ORF Nucleotide The ORF insert of this clone is exactly the same as(RC230814). Sequence: Restriction Sites: SgfI-MluI Cloning Scheme: ACCN: NM_001190996 ORF Size: 1068 bp This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 2 ARPC1A (NM_001190996) Human Tagged ORF Clone – RC230814L4 OTI Disclaimer: The molecular sequence of this clone aligns with the gene accession number as a point of reference only. However, individual transcript sequences of the same gene can differ through naturally occurring variations (e.g. polymorphisms), each with its own valid existence. This clone is substantially in agreement with the reference, but a complete review of all prevailing variants is recommended prior to use. More info OTI Annotation: This clone was engineered to express the complete ORF with an expression tag. Expression varies depending on the nature of the gene. RefSeq: NM_001190996.1, NP_001177925.1 RefSeq ORF: 1071 bp Locus ID: 10552 Protein Pathways: Fc gamma R-mediated phagocytosis, Pathogenic Escherichia coli infection, Regulation of actin cytoskeleton MW: 40.1 kDa Gene Summary: This gene encodes one of seven subunits of the human Arp2/3 protein complex. -

Loss of the Arp2/3 Complex Component ARPC1B Causes Platelet Abnormalities and Predisposes to Inflammatory Disease
ARTICLE Received 9 Nov 2016 | Accepted 31 Jan 2017 | Published 3 Apr 2017 DOI: 10.1038/ncomms14816 OPEN Loss of the Arp2/3 complex component ARPC1B causes platelet abnormalities and predisposes to inflammatory disease Walter H.A. Kahr1,2,3, Fred G. Pluthero1, Abdul Elkadri1,4,5, Neil Warner1,4, Marko Drobac1,3, Chang Hua Chen1,3, Richard W. Lo1,3, Ling Li1, Ren Li1,QiLi1,4, Cornelia Thoeni1,4, Jie Pan1,4, Gabriella Leung1,4, Irene Lara-Corrales6, Ryan Murchie1,4, Ernest Cutz7, Ronald M. Laxer8,9, Julia Upton10, Chaim M. Roifman10, Rae S.M. Yeung1,5,8,11, John H. Brumell1,4,5,12 & Aleixo M. Muise1,3,4,5 Human actin-related protein 2/3 complex (Arp2/3), required for actin filament branching, has two ARPC1 component isoforms, with ARPC1B prominently expressed in blood cells. Here we show in a child with microthrombocytopenia, eosinophilia and inflammatory disease, a homozygous frameshift mutation in ARPC1B (p.Val91Trpfs*30). Platelet lysates reveal no ARPC1B protein and greatly reduced Arp2/3 complex. Missense ARPC1B mutations are identified in an unrelated patient with similar symptoms and ARPC1B deficiency. ARPC1B- deficient platelets are microthrombocytes similar to those seen in Wiskott–Aldrich syndrome that show aberrant spreading consistent with loss of Arp2/3 function. Knockout of ARPC1B in megakaryocytic cells results in decreased proplatelet formation, and as observed in platelets from patients, increased ARPC1A expression. Thus loss of ARPC1B produces a unique set of platelet abnormalities, and is associated with haematopoietic/immune symptoms affecting cell lineages where this isoform predominates. In agreement with recent experimental studies, our findings suggest that ARPC1 isoforms are not functionally interchangeable.