T-Cell Factor 4 Functions As a Tumor Suppressor Whose Disruption Modulates Colon Cell Proliferation and Tumorigenesis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Commentary on WNT7A Implication in Cervical Cancer Development
ndrom Sy es tic & e G n e e n G e f T o Artaza-Irigaray et al., J Genet Syndr Gene Ther 2015, 6:3 Journal of Genetic Syndromes h l e a r n a DOI: 10.4172/2157-7412.1000267 r p u y o J & Gene Therapy ISSN: 2157-7412 Commentary Open Access A Commentary on WNT7A Implication in Cervical Cancer Development Cristina Artaza-Irigaray1,2, Adriana Aguilar-Lemarroy1 and Luis F Jave-Suárez1* 1División de Inmunología, Centro de Investigación Biomédica de Occidente (CIBO), Instituto Mexicano del Seguro Social (IMSS), Guadalajara, Jalisco, Mexico 2Programa de Doctorado en Ciencias Biomédicas, Centro Universitario de Ciencias de la Salud (CUCS) - Universidad de Guadalajara, Jalisco, Mexico Cervical Cancer (CC) is the fourth leading cause of cancer Conversely, silencing Wnt7a in HaCaT cells induced an increase in cell deaths in women worldwide and is associated directly with Human proliferation and migration rates. These results suggest that the loss of papillomavirus (HPV) infection [1]. Many authors have reported Wnt7a expression probably contributes to increased cell proliferation that HPV can immortalize human cells without leading to cell and migration during cervical tumor development. transformation by itself [2,3]. Thus, cervical carcinogenesis is a As responses always lead to new questions, the next step was multistep process involving HPV infection and additional alterations. to elucidate the way in which Wnt7a ligand expression was being In 2005, canonical Wnt signaling pathway activation was proposed repressed. Wnt7a is known to possess tumor suppressor properties as a second hit during epithelial malignant transformation but this in several cancers and is frequently inactivated due to CpG-island hypothesis remains controversial [3,4]. -

Deregulated Wnt/Β-Catenin Program in High-Risk Neuroblastomas Without
Oncogene (2008) 27, 1478–1488 & 2008 Nature Publishing Group All rights reserved 0950-9232/08 $30.00 www.nature.com/onc ONCOGENOMICS Deregulated Wnt/b-catenin program in high-risk neuroblastomas without MYCN amplification X Liu1, P Mazanek1, V Dam1, Q Wang1, H Zhao2, R Guo2, J Jagannathan1, A Cnaan2, JM Maris1,3 and MD Hogarty1,3 1Division of Oncology, The Children’s Hospital of Philadelphia, Philadelphia, PA, USA; 2Department of Biostatistics and Epidemiology, University of Pennsylvania School of Medicine, Philadelphia, PA, USA and 3Department of Pediatrics, University of Pennsylvania School of Medicine, Philadelphia, PA, USA Neuroblastoma (NB) is a frequently lethal tumor of Introduction childhood. MYCN amplification accounts for the aggres- sive phenotype in a subset while the majority have no Neuroblastoma (NB) is a childhood embryonal malig- consistently identified molecular aberration but frequently nancy arising in the peripheral sympathetic nervous express MYC at high levels. We hypothesized that acti- system. Half of all children with NB present with features vated Wnt/b-catenin (CTNNB1) signaling might account that define their tumorsashigh riskwith poor overall for this as MYC is a b-catenin transcriptional target and survival despite intensive therapy (Matthay et al., 1999). multiple embryonal and neural crest malignancies have A subset of these tumors are characterized by high-level oncogenic alterations in this pathway. NB cell lines without genomic amplification of the MYCN proto-oncogene MYCN amplification express higher levels of MYC and (Matthay et al., 1999) but the remainder have no b-catenin (with aberrant nuclear localization) than MYCN- consistently identified aberration to account for their amplified cell lines. -

Estrogen Receptor-<Alpha> Knockout Mice Exhibit Resistance To
Developmental Biology 238, 224–238 (2001) doi:10.1006/dbio.2001.0413, available online at http://www.idealibrary.com on View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Estrogen Receptor-␣ Knockout Mice Exhibit Resistance to the Developmental Effects of Neonatal Diethylstilbestrol Exposure on the Female Reproductive Tract John F. Couse,*,† Darlene Dixon,‡ Mariana Yates,* Alicia B. Moore,‡ Liang Ma,§,1 Richard Maas,§ and Kenneth S. Korach*,2 *Receptor Biology Section, Laboratory of Reproductive and Developmental Toxicology, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, North Carolina 27709; †Department of Environmental and Molecular Toxicology, North Carolina State University, Raleigh, North Carolina 27695; ‡Comparative Pathobiology Section, Laboratory of Experimental Pathology, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, North Carolina 27709; and §Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School and Howard Hughes Medical Institute, Boston, Massachusetts 02115 Data indicate that estrogen-dependent and -independent pathways are involved in the teratogenic/carcinogenic syndrome that follows developmental exposure to 17-estradiol or diethylstilbestrol (DES), a synthetic estrogen. However, the exact role and extent to which each pathway contributes to the resulting pathology remain unknown. We employed the ␣ERKO mouse, which lacks estrogen receptor-␣ (ER␣), to discern the role of ER␣ and estrogen signaling in mediating the effects of neonatal DES exposure. The ␣ERKO provides the potential to expose DES actions mediated by the second known ER, ER, and those that are ER-independent. Wild-type and ␣ERKO females were treated with vehicle or DES (2 g/pup/day for Days 1–5) and terminated after 5 days and 2, 4, 8, 12, and 20 months for biochemical and histomorphological analyses. -

Wnt Proteins Synergize to Activate Β-Catenin Signaling Anshula Alok1, Zhengdeng Lei1,2,*, N
© 2017. Published by The Company of Biologists Ltd | Journal of Cell Science (2017) 130, 1532-1544 doi:10.1242/jcs.198093 RESEARCH ARTICLE Wnt proteins synergize to activate β-catenin signaling Anshula Alok1, Zhengdeng Lei1,2,*, N. Suhas Jagannathan1,2, Simran Kaur1,‡, Nathan Harmston2, Steven G. Rozen1,2, Lisa Tucker-Kellogg1,2 and David M. Virshup1,3,§ ABSTRACT promoters and enhancers to drive expression with distinct Wnt ligands are involved in diverse signaling pathways that are active developmental timing and tissue specificity. However, in both during development, maintenance of tissue homeostasis and in normal and disease states, multiple Wnt genes are often expressed various disease states. While signaling regulated by individual Wnts in combination (Akiri et al., 2009; Bafico et al., 2004; Benhaj et al., has been extensively studied, Wnts are rarely expressed alone, 2006; Suzuki et al., 2004). For example, stromal cells that support the and the consequences of Wnt gene co-expression are not well intestinal stem cell niche express at least six different Wnts at the same understood. Here, we studied the effect of co-expression of Wnts on time (Kabiri et al., 2014). While in isolated instances, specific Wnt β the β-catenin signaling pathway. While some Wnts are deemed ‘non- pairs have been shown to combine to enhance -catenin signaling canonical’ due to their limited ability to activate β-catenin when during embryonic development, whether this is a general expressed alone, unexpectedly, we find that multiple Wnt combinations phenomenon remains unclear (Cha et al., 2008; Cohen et al., 2012; can synergistically activate β-catenin signaling in multiple cell types. -

Estrogen Receptor Alpha (ESR1)-Dependent Regulation of the Mouse Oviductal Transcriptome Katheryn L
University of Kentucky UKnowledge Animal and Food Sciences Faculty Publications Animal and Food Sciences 1-25-2016 Estrogen Receptor Alpha (ESR1)-Dependent Regulation of the Mouse Oviductal Transcriptome Katheryn L. Cerny University of Kentucky, [email protected] Rosanne A. C. Ribeiro University of Kentucky Myoungkun Jeoung University of Kentucky, [email protected] CheMyong Ko University of Illinois at Urbana-Champaign Phillip J. Bridges University of Kentucky, [email protected] Right click to open a feedback form in a new tab to let us know how this document benefits oy u. Follow this and additional works at: https://uknowledge.uky.edu/animalsci_facpub Part of the Animal Sciences Commons, Cell and Developmental Biology Commons, Food Science Commons, and the Physiology Commons Repository Citation Cerny, Katheryn L.; Ribeiro, Rosanne A. C.; Jeoung, Myoungkun; Ko, CheMyong; and Bridges, Phillip J., "Estrogen Receptor Alpha (ESR1)-Dependent Regulation of the Mouse Oviductal Transcriptome" (2016). Animal and Food Sciences Faculty Publications. 7. https://uknowledge.uky.edu/animalsci_facpub/7 This Article is brought to you for free and open access by the Animal and Food Sciences at UKnowledge. It has been accepted for inclusion in Animal and Food Sciences Faculty Publications by an authorized administrator of UKnowledge. For more information, please contact [email protected]. Estrogen Receptor Alpha (ESR1)-Dependent Regulation of the Mouse Oviductal Transcriptome Notes/Citation Information Published in PLOS ONE, v. 11, no. 1, e0147685, p. 1-17. © 2016 Cerny et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. -

Ror2 Functions As a Noncanonical Wnt Receptor That Regulates NMDAR-Mediated Synaptic Transmission
RoR2 functions as a noncanonical Wnt receptor that regulates NMDAR-mediated synaptic transmission Waldo Cerpaa,1, Elena Latorre-Estevesa,b, and Andres Barriaa,2 aDepartment of Physiology and Biophysics and bMolecular and Cellular Biology Program, University of Washington, Seattle, WA 98195 Edited* by Richard L. Huganir, The Johns Hopkins University School of Medicine, Baltimore, MD, and approved March 6, 2015 (received for review September 16, 2014) Wnt signaling has a well-established role as a regulator of nervous receptor for noncanonical Wnt ligands capable of regulating syn- system development, but its role in the maintenance and regula- aptic NMDARs. In hippocampal neurons, activation of RoR2 by tion of established synapses in the mature brain remains poorly noncanonical Wnt ligand Wnt5a activates PKC and JNK, two understood. At excitatory glutamatergic synapses, NMDA receptors kinases involved in the regulation of NMDAR currents. In ad- (NMDARs) have a fundamental role in synaptogenesis, synaptic dition, we show that signaling through RoR2 is necessary for plasticity, and learning and memory; however, it is not known what the maintenance of basal NMDAR-mediated synaptic trans- controls their number and subunit composition. Here we show that mission and the acute regulation of NMDAR synaptic responses the receptor tyrosine kinase-like orphan receptor 2 (RoR2) func- by Wnt5a. tions as a Wnt receptor required to maintain basal NMDAR- Identification of RoR2 as a Wnt receptor that regulates syn- aptic NMDARs provides a mechanism for Wnt signaling to mediated synaptic transmission. In addition, RoR2 activation by a control synaptic transmission and synaptic plasticity acutely, and noncanonical Wnt ligand activates PKC and JNK and acutely en- is a critical first step toward understanding the role played by hances NMDAR synaptic responses. -

Wnt7a Predicts Poor Prognosis, and Contributes to Growth and Metastasis in Tongue Squamous Cell Carcinoma
ONCOLOGY REPORTS 41: 1749-1758, 2019 Wnt7a predicts poor prognosis, and contributes to growth and metastasis in tongue squamous cell carcinoma BO JIA1*, XIAOLING QIU1*, HONGXING CHU1, XIANG SUN1, SHUAIMEI XU2, XINYUAN ZHAO2 and JIANJIANG ZHAO1 Departments of 1Oral Surgery and 2Endodontics, Stomatological Hospital, Southern Medical University, Guangzhou, Guangdong 510280, P.R. China Received May 10, 2018; Accepted September 3, 2018 DOI: 10.3892/or.2019.6974 Abstract. Regional and distant metastases are the principal oncogene, and a potential prognostic and therapeutic target for reasons underlying the high mortality rate associated with patients with TSCC. tongue squamous cell carcinoma (TSCC); however, the precise molecular mechanisms involved in tongue tumorigen- Introduction esis remain unknown. The present study aimed to determine the expression and mechanism of regulation of Wnt7a in the Tongue squamous cell carcinoma (TSCC) is one of the most growth and metastasis of TSCC. Wnt7a mRNA and protein common types of head and neck cancer (HNC) (1,2); >3,000 expression levels were examined in TSCC tissues using reverse people worldwide are diagnosed with TSCC annually and its transcription-quantitative polymerase chain reaction and accounts for 24% of all HNC cases (3). Although many patients immunohistochemical staining. A loss-of-function assay was are identified at the earlier stages of the disease, the 5‑year performed in TSCC cell lines using Wnt7a small interfering survival rate for advanced-stage TSCC is only ~50%, due to RNA or short hairpin RNA, after which, cell proliferation, high invasiveness and metastasis (4). In addition, no effective migration and invasion were analyzed using Cell Counting drugs are currently available for the treatment and control of Kit-8, tumorigenicity and Transwell assays, respectively. -
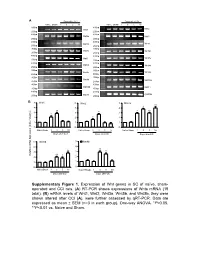
Supplementary Figure 1. Expression of Wnt Genes in SC of Naïve
A Days after CCI Days after CCI Naive Sham 1 3 5 10 Naive Sham 1 3 5 10 500bp 400bp Wnt1 Wnt2 250bp 250bp 400bp 400bp Wnt2b Wnt3 250bp 250bp 400bp 400bp Wnt3a Wnt4 250bp 250bp 300bp 400bp Wnt5a Wnt5b 200bp 250bp 400bp 400bp Wnt6 Wnt7a 250bp 250bp 400bp 400bp Wnt7b Wnt8a 250bp 250bp 400bp 350bp Wnt8b Wnt9a 250bp 200bp 400bp 400bp Wnt9b Wnt10a 250bp 250bp 400bp 400bp Wnt10b Wnt11 250bp 250bp 400bp 400bp Wnt16 GAPDH 250bp 250bp B 5 Wnt1 5 Wnt2 5 Wnt3a ** 4 4 4 * ** ** 3 3 * 3 * * 2 2 * 2 1 1 1 0 0 0 NaïveSham 1 3 5 10 Naïve Sham 1 3 5 10 Naïve Sham 1 3 5 10 Days after CCI Days after CCI Days after CCI 5 Wnt5b 5 Wnt8b 4 4 ** 3 ** 3 ** Relative mRNA Expression (fold of change) ** 2 * 2 1 1 0 0 NaïveSham 1 3 5 10 NaïveSham 1 3 5 10 Days after CCI Days after CCI Supplementary Figure 1 A B Negative Weak Moderate Strong Negative Weak Moderate Strong Large-sized cells 100% 100% 80% 80% 60% 60% 40% 20% 40% 0% 20% Sham CCI-1d CCI-14d Medium-sized cells Wnt3a immunoreactivity cells 0% 100% CGRP(+) IB4(+) 80% Small cells 60% 40% 20% 0% Sham CCI-1d CCI-14d Wnt3a immunoreactivity cells Small cells 100% 80% 60% 40% 20% 0% Sham CCI-1d CCI-14d Fig. S2 4 Fz1 4 Fz3 4 Fz4 4 Fz5 ** 3 3 ** 3 3 2 ** 2 2 ** 2 * 1 1 1 1 0 0 0 0 NaïveSham 1 5 10 NaïveSham 1 5 10 Naïve Sham 1 5 10 NaïveSham 1 5 10 Days after CCI Days after CCI Days after CCI Days after CCI 4 Fz6 4 Fz7 4 Fz8 4 Fz9 (Fold of Change) of (Fold 3 3 3 ** 3 Relative mRNA Expression Expression mRNA Relative ** 2 2 2 ** 2 * 1 1 1 1 0 0 0 0 NaïveSham 1 5 10 NaïveSham 1 5 10 Naïve Sham 1d 5d 10d NaïveSham 1 -

The Role of Wnt Signalling in Excitatory Hippocampal Synapse Formation and Function
The role of Wnt signalling in excitatory hippocampal synapse formation and function Derek Anane UCL PhD Thesis Supervisors: Prof. Patricia C. Salinas Dr. Alasdair Gibb 1 I, Derek Anane confirm that the work presented in this thesis is my own. Where information has been derived from other sources, I confirm that this has been indicated in the thesis. 2 Abstract The formation of a functional neural network is dependent on the correct assembly of cell-cell contacts. Apposition of pre and post synaptic terminals is a highly regulated process culminating in the formation of the functional junction points called synapses. Whilst much is understood of the processes involved in bringing axon and target together less is understood of the mechanisms controlling synapse assembly and maintenance. Wnts are highly glycosylated secretary proteins which have been demonstrated to be involved at several stages of the developing nervous system. Through a range of signalling pathways Wnts are able to produce cellular effects including embryonic patterning, fate and movement. Much recent research has been focused on the role of Wnts in synapse formation and function. In this thesis I present data from hippocampal cultures showing Wnt7a regulation of excitatory synapses. Exposure of developing hippocampal neurons to Wnt7a results in an increase in the density of surface GluA1, GluA2 and GluN1 puncta on dendritic spines. Wnt7a also regulates the co-localisation of postsynaptic glutamate receptor puncta with presynaptic sites labelled with vesicular glutamate transporter protein (vGlut). Interestingly the Wnt7a mediated increase in excitatory synapse formation is no longer present on neurons from mature cultures. At the main postsynaptic site of excitatory synaptic transmission I identified Wnt7a and Dvl mediated maturation of glutamatergic receptor localisation. -

Uterine Epithelial Estrogen Receptor Α Is Dispensable for Proliferation but Essential for Complete Biological and Biochemical Responses
Uterine epithelial estrogen receptor α is dispensable for proliferation but essential for complete biological and biochemical responses Wipawee Winuthayanona, Sylvia C. Hewitta, Grant D. Orvisb,c, Richard R. Behringerc, and Kenneth S. Koracha,1 aLaboratory of Reproductive and Developmental Toxicology, National Institute of Environmental Health Sciences, National Institutes of Health, Department of Health and Human Services, Research Triangle Park, NC 27709; bDevelopmental Biology Program, Sloan–Kettering Institute, York Avenue, New York, NY 10065; and cDepartment of Genetics, University of Texas M.D. Anderson Cancer Center, Houston, TX 77030 Edited by R. Michael Roberts, University of Missouri, Columbia, MO, and approved September 17, 2010 (received for review May 7, 2010) Female fertility requires estrogen to specifically stimulate estrogen uterine epithelial DNA synthesis was stromal ERα-dependent via receptor α (ERα)-dependent growth of the uterine epithelium in induction of autocrine or paracrine mitogenic factors in the stro- adult mice, while immature females show proliferation in both ma that stimulated the epithelial proliferation (5). Evaluating an stroma and epithelium. To address the relative roles of ERα in epithelial-specific role for ERα in tissue proliferation was re- mediating estrogen action in uterine epithelium versus stroma, a cently reported in the mammary gland. Use of a MMTV-Cre uterine epithelial-specific ERα knockout (UtEpiαERKO) mouse line crossed with floxed ERα mice demonstrated that mammary gland was generated by crossing Esr mice with Wnt7a-Cre mice. Expres- growth and ductal epithelial cell proliferation required the mam- sion of Wnt7a directed Cre activity generated selective deletion of mary epithelial ERα (6), which contrasted with previous me- ERα in uterine epithelium, and female UtEpiαERKO are infertile. -

Glyphosate-Based Herbicide Enhances the Uterine Sensitivity to Estradiol in Rats
239 2 Journal of M Guerrero Schimpf et al. Estradiol sensitivity after 239:2 197–213 Endocrinology herbicide exposure RESEARCH Glyphosate-based herbicide enhances the uterine sensitivity to estradiol in rats Marlise Guerrero Schimpf1,2, María M Milesi1,2, Enrique H Luque1,2 and Jorgelina Varayoud1,2 1Instituto de Salud y Ambiente del Litoral (ISAL, UNL-CONICET), Facultad de Bioquímica y Ciencias Biológicas, Universidad Nacional del Litoral, Santa Fe, Argentina 2Cátedra de Fisiología Humana, Facultad de Bioquímica y Ciencias Biológicas, Universidad Nacional del Litoral, Santa Fe, Argentina Correspondence should be addressed to J Varayoud: [email protected] Abstract In a previous work, we detected that postnatal exposure to a glyphosate-based Key Words herbicide (GBH) alters uterine development in prepubertal rats causing endometrial f estrogen receptors hyperplasia and increasing cell proliferation. Our goal was to determine whether f uterus exposure to low dose of a GBH during postnatal development might enhance f neonatal the sensitivity of the uterus to an estrogenic treatment. Female Wistar pups were f rat subcutaneously injected with saline solution (control) or GBH using the reference dose (2 mg/kg/day, EPA) on postnatal days (PND) 1, 3, 5 and 7. At weaning (PND21), female rats were bilaterally ovariectomized and treated with silastic capsules containing 17β-estradiol (E2, 1 mg/mL) until they were 2 months of age. On PND60, uterine samples were removed and processed for histology, immunohistochemistry and mRNA extraction to evaluate: (i) uterine morphology, (ii) uterine cell proliferation by the detection of Ki67, (iii) the expression of the estrogen receptors alpha (ESR1) and beta (ESR2) and (iv) the expression of WNT7A and CTNNB1. -

Canonical Wnt Signaling Is Necessary for Object Recognition Memory Consolidation
The Journal of Neuroscience, July 31, 2013 • 33(31):12619–12626 • 12619 Behavioral/Cognitive Canonical Wnt Signaling is Necessary for Object Recognition Memory Consolidation Ashley M. Fortress, Sarah L. Schram, Jennifer J. Tuscher, and Karyn M. Frick Department of Psychology, University of Wisconsin-Milwaukee, Milwaukee, Wisconsin 53211 Wnt signaling has emerged as a potent regulator of hippocampal synaptic function, although no evidence yet supports a critical role for Wnt signaling in hippocampal memory. Here, we sought to determine whether canonical -catenin-dependent Wnt signaling is neces- sary for hippocampal memory consolidation. Immediately after training in a hippocampal-dependent object recognition task, mice received a dorsal hippocampal (DH) infusion of vehicle or the canonical Wnt antagonist Dickkopf-1 (Dkk-1; 50, 100, or 200 ng/hemi- sphere). Twenty-four hours later, mice receiving vehicle remembered the familiar object explored during training. However, mice receiving Dkk-1 exhibited no memory for the training object, indicating that object recognition memory consolidation is dependent on canonical Wnt signaling. To determine how Dkk-1 affects canonical Wnt signaling, mice were infused with vehicle or 50 ng/hemisphere Dkk-1 and protein levels of Wnt-related proteins (Dkk-1, GSK3, -catenin, TCF1, LEF1, Cyclin D1, c-myc, Wnt7a, Wnt1, and PSD95) were measured in the dorsal hippocampus 5 min or 4 h later. Dkk-1 produced a rapid increase in Dkk-1 protein levels and a decrease in phosphorylated GSK3 levels, followed by a decrease in -catenin, TCF1, LEF1, Cyclin D1, c-myc, Wnt7a, and PSD95 protein levels 4 h later. These data suggest that alterations in Wnt/GSK3/-catenin signaling may underlie the memory impairments induced by Dkk-1.