Estrogen Receptor Alpha (ESR1)-Dependent Regulation of the Mouse Oviductal Transcriptome Katheryn L
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genetics of Interleukin 1 Receptor-Like 1 in Immune and Inflammatory Diseases
Current Genomics, 2010, 11, 591-606 591 Genetics of Interleukin 1 Receptor-Like 1 in Immune and Inflammatory Diseases Loubna Akhabir and Andrew Sandford* Department of Medicine, University of British Columbia, UBC James Hogg Research Centre, Providence Heart + Lung Institute, Room 166, St. Paul's Hospital, 1081 Burrard Street, Vancouver, BC V6Z 1Y6, Canada Abstract: Interleukin 1 receptor-like 1 (IL1RL1) is gaining in recognition due to its involvement in immune/inflamma- tory disorders. Well-designed animal studies have shown its critical role in experimental allergic inflammation and human in vitro studies have consistently demonstrated its up-regulation in several conditions such as asthma and rheumatoid ar- thritis. The ligand for IL1RL1 is IL33 which emerged as playing an important role in initiating eosinophilic inflammation and activating other immune cells resulting in an allergic phenotype. An IL1RL1 single nucleotide polymorphism (SNP) was among the most significant results of a genome-wide scan inves- tigating eosinophil counts; in the same study, this SNP associated with asthma in 10 populations. The IL1RL1 gene resides in a region of high linkage disequilibrium containing interleukin 1 receptor genes as well as in- terleukin 18 receptor and accessory genes. This poses a challenge to researchers interested in deciphering genetic associa- tion signals in the region as all of the genes represent interesting candidates for asthma and allergic disease. The IL1RL1 gene and its resulting soluble and receptor proteins have emerged as key regulators of the inflammatory proc- ess implicated in a large variety of human pathologies We review the function and expression of the IL1RL1 gene. -

A Commentary on WNT7A Implication in Cervical Cancer Development
ndrom Sy es tic & e G n e e n G e f T o Artaza-Irigaray et al., J Genet Syndr Gene Ther 2015, 6:3 Journal of Genetic Syndromes h l e a r n a DOI: 10.4172/2157-7412.1000267 r p u y o J & Gene Therapy ISSN: 2157-7412 Commentary Open Access A Commentary on WNT7A Implication in Cervical Cancer Development Cristina Artaza-Irigaray1,2, Adriana Aguilar-Lemarroy1 and Luis F Jave-Suárez1* 1División de Inmunología, Centro de Investigación Biomédica de Occidente (CIBO), Instituto Mexicano del Seguro Social (IMSS), Guadalajara, Jalisco, Mexico 2Programa de Doctorado en Ciencias Biomédicas, Centro Universitario de Ciencias de la Salud (CUCS) - Universidad de Guadalajara, Jalisco, Mexico Cervical Cancer (CC) is the fourth leading cause of cancer Conversely, silencing Wnt7a in HaCaT cells induced an increase in cell deaths in women worldwide and is associated directly with Human proliferation and migration rates. These results suggest that the loss of papillomavirus (HPV) infection [1]. Many authors have reported Wnt7a expression probably contributes to increased cell proliferation that HPV can immortalize human cells without leading to cell and migration during cervical tumor development. transformation by itself [2,3]. Thus, cervical carcinogenesis is a As responses always lead to new questions, the next step was multistep process involving HPV infection and additional alterations. to elucidate the way in which Wnt7a ligand expression was being In 2005, canonical Wnt signaling pathway activation was proposed repressed. Wnt7a is known to possess tumor suppressor properties as a second hit during epithelial malignant transformation but this in several cancers and is frequently inactivated due to CpG-island hypothesis remains controversial [3,4]. -

How Relevant Are Bone Marrow-Derived Mast Cells (Bmmcs) As Models for Tissue Mast Cells? a Comparative Transcriptome Analysis of Bmmcs and Peritoneal Mast Cells
cells Article How Relevant Are Bone Marrow-Derived Mast Cells (BMMCs) as Models for Tissue Mast Cells? A Comparative Transcriptome Analysis of BMMCs and Peritoneal Mast Cells 1, 2, 1 1 2,3 Srinivas Akula y , Aida Paivandy y, Zhirong Fu , Michael Thorpe , Gunnar Pejler and Lars Hellman 1,* 1 Department of Cell and Molecular Biology, Uppsala University, The Biomedical Center, Box 596, SE-751 24 Uppsala, Sweden; [email protected] (S.A.); [email protected] (Z.F.); [email protected] (M.T.) 2 Department of Medical Biochemistry and Microbiology, Uppsala University, The Biomedical Center, Box 589, SE-751 23 Uppsala, Sweden; [email protected] (A.P.); [email protected] (G.P.) 3 Department of Anatomy, Physiology and Biochemistry, Swedish University of Agricultural Sciences, Box 7011, SE-75007 Uppsala, Sweden * Correspondence: [email protected]; Tel.: +46-(0)18-471-4532; Fax: +46-(0)18-471-4862 These authors contributed equally to this work. y Received: 29 July 2020; Accepted: 16 September 2020; Published: 17 September 2020 Abstract: Bone marrow-derived mast cells (BMMCs) are often used as a model system for studies of the role of MCs in health and disease. These cells are relatively easy to obtain from total bone marrow cells by culturing under the influence of IL-3 or stem cell factor (SCF). After 3 to 4 weeks in culture, a nearly homogenous cell population of toluidine blue-positive cells are often obtained. However, the question is how relevant equivalents these cells are to normal tissue MCs. By comparing the total transcriptome of purified peritoneal MCs with BMMCs, here we obtained a comparative view of these cells. -

The Interleukin-18 Receptor Is Differentially Expressed in Whole
1 The interleukin-18 receptor is differentially expressed in the blood during sepsis. 2 3 Shahan Mamoor, MS1 4 [email protected] East Islip, NY, USA 5 6 We probed published and public microarray datasets1,2 to discover the most significant gene expression changes in the blood of patients with sepsis. We found significant induction 7 expression of IL18RAP and IL18R1, genes encoding subunits of the interleukin-18 receptor, in 8 whole blood from patients with sepsis. 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 Keywords: sepsis, septic shock, interleukin-18, IL18R1, IL18RAP, systems biology of septic 26 shock. 27 28 PAGE 1 OF 13 1 Septic shock is a leading cause of mortality in the United States and worldwide3. We 2 used published and public microarray datasets1,2 to identify differentially expressed genes in the 3 4 blood of patients with sepsis. We identified IL18R1 and IL18RAP as among the genes most 5 differentially expressed in blood in the septic state. 6 7 Methods 8 9 We utilized microarray datasets GSE1001591 and GSE264402 for this differential gene 10 11 expression analysis of blood cells during sepsis. GSE100509 was generated with whole blood 12 using Illumina HumanWG-6 v3.0 expression beadchip technology with n=12 whole blood from 13 control subjects and n=33 whole blood from sepsis patients. GSE26440 was generated using 14 15 Affymetrix Human Genome U133 Plus 2.0 Array technology with n=32 control subjects and 16 n=98 sepsis patients. The Benjamini and Hochberg method of p-value adjustment was used for 17 ranking of differential expression but raw p-values were used for assessment of statistical 18 19 significance of global differential expression. -

Deregulated Wnt/Β-Catenin Program in High-Risk Neuroblastomas Without
Oncogene (2008) 27, 1478–1488 & 2008 Nature Publishing Group All rights reserved 0950-9232/08 $30.00 www.nature.com/onc ONCOGENOMICS Deregulated Wnt/b-catenin program in high-risk neuroblastomas without MYCN amplification X Liu1, P Mazanek1, V Dam1, Q Wang1, H Zhao2, R Guo2, J Jagannathan1, A Cnaan2, JM Maris1,3 and MD Hogarty1,3 1Division of Oncology, The Children’s Hospital of Philadelphia, Philadelphia, PA, USA; 2Department of Biostatistics and Epidemiology, University of Pennsylvania School of Medicine, Philadelphia, PA, USA and 3Department of Pediatrics, University of Pennsylvania School of Medicine, Philadelphia, PA, USA Neuroblastoma (NB) is a frequently lethal tumor of Introduction childhood. MYCN amplification accounts for the aggres- sive phenotype in a subset while the majority have no Neuroblastoma (NB) is a childhood embryonal malig- consistently identified molecular aberration but frequently nancy arising in the peripheral sympathetic nervous express MYC at high levels. We hypothesized that acti- system. Half of all children with NB present with features vated Wnt/b-catenin (CTNNB1) signaling might account that define their tumorsashigh riskwith poor overall for this as MYC is a b-catenin transcriptional target and survival despite intensive therapy (Matthay et al., 1999). multiple embryonal and neural crest malignancies have A subset of these tumors are characterized by high-level oncogenic alterations in this pathway. NB cell lines without genomic amplification of the MYCN proto-oncogene MYCN amplification express higher levels of MYC and (Matthay et al., 1999) but the remainder have no b-catenin (with aberrant nuclear localization) than MYCN- consistently identified aberration to account for their amplified cell lines. -

Estrogen Receptor-<Alpha> Knockout Mice Exhibit Resistance To
Developmental Biology 238, 224–238 (2001) doi:10.1006/dbio.2001.0413, available online at http://www.idealibrary.com on View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Estrogen Receptor-␣ Knockout Mice Exhibit Resistance to the Developmental Effects of Neonatal Diethylstilbestrol Exposure on the Female Reproductive Tract John F. Couse,*,† Darlene Dixon,‡ Mariana Yates,* Alicia B. Moore,‡ Liang Ma,§,1 Richard Maas,§ and Kenneth S. Korach*,2 *Receptor Biology Section, Laboratory of Reproductive and Developmental Toxicology, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, North Carolina 27709; †Department of Environmental and Molecular Toxicology, North Carolina State University, Raleigh, North Carolina 27695; ‡Comparative Pathobiology Section, Laboratory of Experimental Pathology, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, North Carolina 27709; and §Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School and Howard Hughes Medical Institute, Boston, Massachusetts 02115 Data indicate that estrogen-dependent and -independent pathways are involved in the teratogenic/carcinogenic syndrome that follows developmental exposure to 17-estradiol or diethylstilbestrol (DES), a synthetic estrogen. However, the exact role and extent to which each pathway contributes to the resulting pathology remain unknown. We employed the ␣ERKO mouse, which lacks estrogen receptor-␣ (ER␣), to discern the role of ER␣ and estrogen signaling in mediating the effects of neonatal DES exposure. The ␣ERKO provides the potential to expose DES actions mediated by the second known ER, ER, and those that are ER-independent. Wild-type and ␣ERKO females were treated with vehicle or DES (2 g/pup/day for Days 1–5) and terminated after 5 days and 2, 4, 8, 12, and 20 months for biochemical and histomorphological analyses. -

Wnt Proteins Synergize to Activate Β-Catenin Signaling Anshula Alok1, Zhengdeng Lei1,2,*, N
© 2017. Published by The Company of Biologists Ltd | Journal of Cell Science (2017) 130, 1532-1544 doi:10.1242/jcs.198093 RESEARCH ARTICLE Wnt proteins synergize to activate β-catenin signaling Anshula Alok1, Zhengdeng Lei1,2,*, N. Suhas Jagannathan1,2, Simran Kaur1,‡, Nathan Harmston2, Steven G. Rozen1,2, Lisa Tucker-Kellogg1,2 and David M. Virshup1,3,§ ABSTRACT promoters and enhancers to drive expression with distinct Wnt ligands are involved in diverse signaling pathways that are active developmental timing and tissue specificity. However, in both during development, maintenance of tissue homeostasis and in normal and disease states, multiple Wnt genes are often expressed various disease states. While signaling regulated by individual Wnts in combination (Akiri et al., 2009; Bafico et al., 2004; Benhaj et al., has been extensively studied, Wnts are rarely expressed alone, 2006; Suzuki et al., 2004). For example, stromal cells that support the and the consequences of Wnt gene co-expression are not well intestinal stem cell niche express at least six different Wnts at the same understood. Here, we studied the effect of co-expression of Wnts on time (Kabiri et al., 2014). While in isolated instances, specific Wnt β the β-catenin signaling pathway. While some Wnts are deemed ‘non- pairs have been shown to combine to enhance -catenin signaling canonical’ due to their limited ability to activate β-catenin when during embryonic development, whether this is a general expressed alone, unexpectedly, we find that multiple Wnt combinations phenomenon remains unclear (Cha et al., 2008; Cohen et al., 2012; can synergistically activate β-catenin signaling in multiple cell types. -

Ror2 Functions As a Noncanonical Wnt Receptor That Regulates NMDAR-Mediated Synaptic Transmission
RoR2 functions as a noncanonical Wnt receptor that regulates NMDAR-mediated synaptic transmission Waldo Cerpaa,1, Elena Latorre-Estevesa,b, and Andres Barriaa,2 aDepartment of Physiology and Biophysics and bMolecular and Cellular Biology Program, University of Washington, Seattle, WA 98195 Edited* by Richard L. Huganir, The Johns Hopkins University School of Medicine, Baltimore, MD, and approved March 6, 2015 (received for review September 16, 2014) Wnt signaling has a well-established role as a regulator of nervous receptor for noncanonical Wnt ligands capable of regulating syn- system development, but its role in the maintenance and regula- aptic NMDARs. In hippocampal neurons, activation of RoR2 by tion of established synapses in the mature brain remains poorly noncanonical Wnt ligand Wnt5a activates PKC and JNK, two understood. At excitatory glutamatergic synapses, NMDA receptors kinases involved in the regulation of NMDAR currents. In ad- (NMDARs) have a fundamental role in synaptogenesis, synaptic dition, we show that signaling through RoR2 is necessary for plasticity, and learning and memory; however, it is not known what the maintenance of basal NMDAR-mediated synaptic trans- controls their number and subunit composition. Here we show that mission and the acute regulation of NMDAR synaptic responses the receptor tyrosine kinase-like orphan receptor 2 (RoR2) func- by Wnt5a. tions as a Wnt receptor required to maintain basal NMDAR- Identification of RoR2 as a Wnt receptor that regulates syn- aptic NMDARs provides a mechanism for Wnt signaling to mediated synaptic transmission. In addition, RoR2 activation by a control synaptic transmission and synaptic plasticity acutely, and noncanonical Wnt ligand activates PKC and JNK and acutely en- is a critical first step toward understanding the role played by hances NMDAR synaptic responses. -

Wnt7a Predicts Poor Prognosis, and Contributes to Growth and Metastasis in Tongue Squamous Cell Carcinoma
ONCOLOGY REPORTS 41: 1749-1758, 2019 Wnt7a predicts poor prognosis, and contributes to growth and metastasis in tongue squamous cell carcinoma BO JIA1*, XIAOLING QIU1*, HONGXING CHU1, XIANG SUN1, SHUAIMEI XU2, XINYUAN ZHAO2 and JIANJIANG ZHAO1 Departments of 1Oral Surgery and 2Endodontics, Stomatological Hospital, Southern Medical University, Guangzhou, Guangdong 510280, P.R. China Received May 10, 2018; Accepted September 3, 2018 DOI: 10.3892/or.2019.6974 Abstract. Regional and distant metastases are the principal oncogene, and a potential prognostic and therapeutic target for reasons underlying the high mortality rate associated with patients with TSCC. tongue squamous cell carcinoma (TSCC); however, the precise molecular mechanisms involved in tongue tumorigen- Introduction esis remain unknown. The present study aimed to determine the expression and mechanism of regulation of Wnt7a in the Tongue squamous cell carcinoma (TSCC) is one of the most growth and metastasis of TSCC. Wnt7a mRNA and protein common types of head and neck cancer (HNC) (1,2); >3,000 expression levels were examined in TSCC tissues using reverse people worldwide are diagnosed with TSCC annually and its transcription-quantitative polymerase chain reaction and accounts for 24% of all HNC cases (3). Although many patients immunohistochemical staining. A loss-of-function assay was are identified at the earlier stages of the disease, the 5‑year performed in TSCC cell lines using Wnt7a small interfering survival rate for advanced-stage TSCC is only ~50%, due to RNA or short hairpin RNA, after which, cell proliferation, high invasiveness and metastasis (4). In addition, no effective migration and invasion were analyzed using Cell Counting drugs are currently available for the treatment and control of Kit-8, tumorigenicity and Transwell assays, respectively. -

Triangulating Molecular Evidence to Prioritise Candidate Causal Genes at Established Atopic Dermatitis Loci
medRxiv preprint doi: https://doi.org/10.1101/2020.11.30.20240838; this version posted November 30, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY-ND 4.0 International license . Triangulating molecular evidence to prioritise candidate causal genes at established atopic dermatitis loci Maria K Sobczyk1, Tom G Richardson1, Verena Zuber2,3, Josine L Min1, eQTLGen Consortium4, BIOS Consortium5, GoDMC, Tom R Gaunt1, Lavinia Paternoster1* 1) MRC Integrative Epidemiology Unit, Bristol Medical School, University of Bristol, Bristol, UK 2) Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, London, UK 3) MRC Biostatistics Unit, School of Clinical Medicine, University of Cambridge, Cambridge, UK 4) Members of the eQTLGen Consortium are listed in: Supplementary_Consortium_members.docx 5) Members of the BIOS Consortium are listed in: Supplementary_Consortium_members.docx Abstract Background: Genome-wide association studies for atopic dermatitis (AD, eczema) have identified 25 reproducible loci associated in populations of European descent. We attempt to prioritise candidate causal genes at these loci using a multifaceted bioinformatic approach and extensive molecular resources compiled into a novel pipeline: ADGAPP (Atopic Dermatitis GWAS Annotation & Prioritisation Pipeline). Methods: We identified a comprehensive list of 103 accessible -
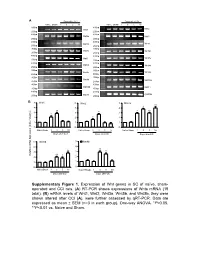
Supplementary Figure 1. Expression of Wnt Genes in SC of Naïve
A Days after CCI Days after CCI Naive Sham 1 3 5 10 Naive Sham 1 3 5 10 500bp 400bp Wnt1 Wnt2 250bp 250bp 400bp 400bp Wnt2b Wnt3 250bp 250bp 400bp 400bp Wnt3a Wnt4 250bp 250bp 300bp 400bp Wnt5a Wnt5b 200bp 250bp 400bp 400bp Wnt6 Wnt7a 250bp 250bp 400bp 400bp Wnt7b Wnt8a 250bp 250bp 400bp 350bp Wnt8b Wnt9a 250bp 200bp 400bp 400bp Wnt9b Wnt10a 250bp 250bp 400bp 400bp Wnt10b Wnt11 250bp 250bp 400bp 400bp Wnt16 GAPDH 250bp 250bp B 5 Wnt1 5 Wnt2 5 Wnt3a ** 4 4 4 * ** ** 3 3 * 3 * * 2 2 * 2 1 1 1 0 0 0 NaïveSham 1 3 5 10 Naïve Sham 1 3 5 10 Naïve Sham 1 3 5 10 Days after CCI Days after CCI Days after CCI 5 Wnt5b 5 Wnt8b 4 4 ** 3 ** 3 ** Relative mRNA Expression (fold of change) ** 2 * 2 1 1 0 0 NaïveSham 1 3 5 10 NaïveSham 1 3 5 10 Days after CCI Days after CCI Supplementary Figure 1 A B Negative Weak Moderate Strong Negative Weak Moderate Strong Large-sized cells 100% 100% 80% 80% 60% 60% 40% 20% 40% 0% 20% Sham CCI-1d CCI-14d Medium-sized cells Wnt3a immunoreactivity cells 0% 100% CGRP(+) IB4(+) 80% Small cells 60% 40% 20% 0% Sham CCI-1d CCI-14d Wnt3a immunoreactivity cells Small cells 100% 80% 60% 40% 20% 0% Sham CCI-1d CCI-14d Fig. S2 4 Fz1 4 Fz3 4 Fz4 4 Fz5 ** 3 3 ** 3 3 2 ** 2 2 ** 2 * 1 1 1 1 0 0 0 0 NaïveSham 1 5 10 NaïveSham 1 5 10 Naïve Sham 1 5 10 NaïveSham 1 5 10 Days after CCI Days after CCI Days after CCI Days after CCI 4 Fz6 4 Fz7 4 Fz8 4 Fz9 (Fold of Change) of (Fold 3 3 3 ** 3 Relative mRNA Expression Expression mRNA Relative ** 2 2 2 ** 2 * 1 1 1 1 0 0 0 0 NaïveSham 1 5 10 NaïveSham 1 5 10 Naïve Sham 1d 5d 10d NaïveSham 1 -

The Role of Wnt Signalling in Excitatory Hippocampal Synapse Formation and Function
The role of Wnt signalling in excitatory hippocampal synapse formation and function Derek Anane UCL PhD Thesis Supervisors: Prof. Patricia C. Salinas Dr. Alasdair Gibb 1 I, Derek Anane confirm that the work presented in this thesis is my own. Where information has been derived from other sources, I confirm that this has been indicated in the thesis. 2 Abstract The formation of a functional neural network is dependent on the correct assembly of cell-cell contacts. Apposition of pre and post synaptic terminals is a highly regulated process culminating in the formation of the functional junction points called synapses. Whilst much is understood of the processes involved in bringing axon and target together less is understood of the mechanisms controlling synapse assembly and maintenance. Wnts are highly glycosylated secretary proteins which have been demonstrated to be involved at several stages of the developing nervous system. Through a range of signalling pathways Wnts are able to produce cellular effects including embryonic patterning, fate and movement. Much recent research has been focused on the role of Wnts in synapse formation and function. In this thesis I present data from hippocampal cultures showing Wnt7a regulation of excitatory synapses. Exposure of developing hippocampal neurons to Wnt7a results in an increase in the density of surface GluA1, GluA2 and GluN1 puncta on dendritic spines. Wnt7a also regulates the co-localisation of postsynaptic glutamate receptor puncta with presynaptic sites labelled with vesicular glutamate transporter protein (vGlut). Interestingly the Wnt7a mediated increase in excitatory synapse formation is no longer present on neurons from mature cultures. At the main postsynaptic site of excitatory synaptic transmission I identified Wnt7a and Dvl mediated maturation of glutamatergic receptor localisation.