Resolution of Ray-Finned Fish Phylogeny and Timing Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Critical Habitat for Canterbury Freshwater Fish, Kōura/Kēkēwai and Kākahi
CRITICAL HABITAT FOR CANTERBURY FRESHWATER FISH, KŌURA/KĒKĒWAI AND KĀKAHI REPORT PREPARED FOR CANTERBURY REGIONAL COUNCIL BY RICHARD ALLIBONE WATERWAYS CONSULTING REPORT NUMBER: 55-2018 AND DUNCAN GRAY CANTERBURY REGIONAL COUNCIL DATE: DECEMBER 2018 EXECUTIVE SUMMARY Aquatic habitat in Canterbury supports a range of native freshwater fish and the mega macroinvertebrates kōura/kēkēwai (crayfish) and kākahi (mussel). Loss of habitat, barriers to fish passage, water quality and water quantity issues present management challenges when we seek to protect this freshwater fauna while providing for human use. Water plans in Canterbury are intended to set rules for the use of water, the quality of water in aquatic systems and activities that occur within and adjacent to aquatic areas. To inform the planning and resource consent processes, information on the distribution of species and their critical habitat requirements can be used to provide for their protection. This report assesses the conservation status and distributions of indigenous freshwater fish, kēkēwai and kākahi in the Canterbury region. The report identifies the geographic distribution of these species and provides information on the critical habitat requirements of these species and/or populations. Water Ways Consulting Ltd Critical habitats for Canterbury aquatic fauna Table of Contents 1 Introduction ......................................................................................................................................... 1 2 Methods .............................................................................................................................................. -

Field Guide to the Nonindigenous Marine Fishes of Florida
Field Guide to the Nonindigenous Marine Fishes of Florida Schofield, P. J., J. A. Morris, Jr. and L. Akins Mention of trade names or commercial products does not constitute endorsement or recommendation for their use by the United States goverment. Pamela J. Schofield, Ph.D. U.S. Geological Survey Florida Integrated Science Center 7920 NW 71st Street Gainesville, FL 32653 [email protected] James A. Morris, Jr., Ph.D. National Oceanic and Atmospheric Administration National Ocean Service National Centers for Coastal Ocean Science Center for Coastal Fisheries and Habitat Research 101 Pivers Island Road Beaufort, NC 28516 [email protected] Lad Akins Reef Environmental Education Foundation (REEF) 98300 Overseas Highway Key Largo, FL 33037 [email protected] Suggested Citation: Schofield, P. J., J. A. Morris, Jr. and L. Akins. 2009. Field Guide to Nonindigenous Marine Fishes of Florida. NOAA Technical Memorandum NOS NCCOS 92. Field Guide to Nonindigenous Marine Fishes of Florida Pamela J. Schofield, Ph.D. James A. Morris, Jr., Ph.D. Lad Akins NOAA, National Ocean Service National Centers for Coastal Ocean Science NOAA Technical Memorandum NOS NCCOS 92. September 2009 United States Department of National Oceanic and National Ocean Service Commerce Atmospheric Administration Gary F. Locke Jane Lubchenco John H. Dunnigan Secretary Administrator Assistant Administrator Table of Contents Introduction ................................................................................................ i Methods .....................................................................................................ii -

A Global Assessment of Parasite Diversity in Galaxiid Fishes
diversity Article A Global Assessment of Parasite Diversity in Galaxiid Fishes Rachel A. Paterson 1,*, Gustavo P. Viozzi 2, Carlos A. Rauque 2, Verónica R. Flores 2 and Robert Poulin 3 1 The Norwegian Institute for Nature Research, P.O. Box 5685, Torgarden, 7485 Trondheim, Norway 2 Laboratorio de Parasitología, INIBIOMA, CONICET—Universidad Nacional del Comahue, Quintral 1250, San Carlos de Bariloche 8400, Argentina; [email protected] (G.P.V.); [email protected] (C.A.R.); veronicaroxanafl[email protected] (V.R.F.) 3 Department of Zoology, University of Otago, P.O. Box 56, Dunedin 9054, New Zealand; [email protected] * Correspondence: [email protected]; Tel.: +47-481-37-867 Abstract: Free-living species often receive greater conservation attention than the parasites they support, with parasite conservation often being hindered by a lack of parasite biodiversity knowl- edge. This study aimed to determine the current state of knowledge regarding parasites of the Southern Hemisphere freshwater fish family Galaxiidae, in order to identify knowledge gaps to focus future research attention. Specifically, we assessed how galaxiid–parasite knowledge differs among geographic regions in relation to research effort (i.e., number of studies or fish individuals examined, extent of tissue examination, taxonomic resolution), in addition to ecological traits known to influ- ence parasite richness. To date, ~50% of galaxiid species have been examined for parasites, though the majority of studies have focused on single parasite taxa rather than assessing the full diversity of macro- and microparasites. The highest number of parasites were observed from Argentinean galaxiids, and studies in all geographic regions were biased towards the highly abundant and most widely distributed galaxiid species, Galaxias maculatus. -

Phylogeny Classification Additional Readings Clupeomorpha and Ostariophysi
Teleostei - AccessScience from McGraw-Hill Education http://www.accessscience.com/content/teleostei/680400 (http://www.accessscience.com/) Article by: Boschung, Herbert Department of Biological Sciences, University of Alabama, Tuscaloosa, Alabama. Gardiner, Brian Linnean Society of London, Burlington House, Piccadilly, London, United Kingdom. Publication year: 2014 DOI: http://dx.doi.org/10.1036/1097-8542.680400 (http://dx.doi.org/10.1036/1097-8542.680400) Content Morphology Euteleostei Bibliography Phylogeny Classification Additional Readings Clupeomorpha and Ostariophysi The most recent group of actinopterygians (rayfin fishes), first appearing in the Upper Triassic (Fig. 1). About 26,840 species are contained within the Teleostei, accounting for more than half of all living vertebrates and over 96% of all living fishes. Teleosts comprise 517 families, of which 69 are extinct, leaving 448 extant families; of these, about 43% have no fossil record. See also: Actinopterygii (/content/actinopterygii/009100); Osteichthyes (/content/osteichthyes/478500) Fig. 1 Cladogram showing the relationships of the extant teleosts with the other extant actinopterygians. (J. S. Nelson, Fishes of the World, 4th ed., Wiley, New York, 2006) 1 of 9 10/7/2015 1:07 PM Teleostei - AccessScience from McGraw-Hill Education http://www.accessscience.com/content/teleostei/680400 Morphology Much of the evidence for teleost monophyly (evolving from a common ancestral form) and relationships comes from the caudal skeleton and concomitant acquisition of a homocercal tail (upper and lower lobes of the caudal fin are symmetrical). This type of tail primitively results from an ontogenetic fusion of centra (bodies of vertebrae) and the possession of paired bracing bones located bilaterally along the dorsal region of the caudal skeleton, derived ontogenetically from the neural arches (uroneurals) of the ural (tail) centra. -

New Insights on the Sister Lineage of Percomorph Fishes with an Anchored Hybrid Enrichment Dataset
Molecular Phylogenetics and Evolution 110 (2017) 27–38 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev New insights on the sister lineage of percomorph fishes with an anchored hybrid enrichment dataset ⇑ Alex Dornburg a, , Jeffrey P. Townsend b,c,d, Willa Brooks a, Elizabeth Spriggs b, Ron I. Eytan e, Jon A. Moore f,g, Peter C. Wainwright h, Alan Lemmon i, Emily Moriarty Lemmon j, Thomas J. Near b,k a North Carolina Museum of Natural Sciences, Raleigh, NC, USA b Department of Ecology & Evolutionary Biology and Peabody Museum of Natural History, Yale University, New Haven, CT 06520, USA c Program in Computational Biology and Bioinformatics, Yale University, New Haven, CT 06520, USA d Department of Biostatistics, Yale University, New Haven, CT 06510, USA e Marine Biology Department, Texas A&M University at Galveston, Galveston, TX 77554, USA f Florida Atlantic University, Wilkes Honors College, Jupiter, FL 33458, USA g Florida Atlantic University, Harbor Branch Oceanographic Institution, Fort Pierce, FL 34946, USA h Department of Evolution & Ecology, University of California, Davis, CA 95616, USA i Department of Scientific Computing, Florida State University, 400 Dirac Science Library, Tallahassee, FL 32306, USA j Department of Biological Science, Florida State University, 319 Stadium Drive, Tallahassee, FL 32306, USA k Peabody Museum of Natural History, Yale University, New Haven, CT 06520, USA article info abstract Article history: Percomorph fishes represent over 17,100 species, including several model organisms and species of eco- Received 12 April 2016 nomic importance. Despite continuous advances in the resolution of the percomorph Tree of Life, resolu- Revised 22 February 2017 tion of the sister lineage to Percomorpha remains inconsistent but restricted to a small number of Accepted 25 February 2017 candidate lineages. -

1 Exon Probe Sets and Bioinformatics Pipelines for All Levels of Fish Phylogenomics
bioRxiv preprint doi: https://doi.org/10.1101/2020.02.18.949735; this version posted February 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Exon probe sets and bioinformatics pipelines for all levels of fish phylogenomics 2 3 Lily C. Hughes1,2,3,*, Guillermo Ortí1,3, Hadeel Saad1, Chenhong Li4, William T. White5, Carole 4 C. Baldwin3, Keith A. Crandall1,2, Dahiana Arcila3,6,7, and Ricardo Betancur-R.7 5 6 1 Department of Biological Sciences, George Washington University, Washington, D.C., U.S.A. 7 2 Computational Biology Institute, Milken Institute of Public Health, George Washington 8 University, Washington, D.C., U.S.A. 9 3 Department of Vertebrate Zoology, National Museum of Natural History, Smithsonian 10 Institution, Washington, D.C., U.S.A. 11 4 College of Fisheries and Life Sciences, Shanghai Ocean University, Shanghai, China 12 5 CSIRO Australian National Fish Collection, National Research Collections of Australia, 13 Hobart, TAS, Australia 14 6 Sam Noble Oklahoma Museum of Natural History, Norman, O.K., U.S.A. 15 7 Department of Biology, University of Oklahoma, Norman, O.K., U.S.A. 16 17 *Corresponding author: Lily C. Hughes, [email protected]. 18 Current address: Department of Organismal Biology and Anatomy, University of Chicago, 19 Chicago, IL. 20 21 Keywords: Actinopterygii, Protein coding, Systematics, Phylogenetics, Evolution, Target 22 capture 23 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.02.18.949735; this version posted February 19, 2020. -

Vertebrate Proteins Predicted from Genomic Sequences
Vertebrate proteins predicted from genomic sequences VWD C8 TIL PTS Mucin2_WxxW F5_F8_type_C FCGBP_N VWC Lethenteron_camtschaticum Cyclostomata; Hyperoartia; Petromyzontiformes; Petromyzontidae; Lethenteron Lethenteron_camtschaticum.0.pep1 Petromyzon_marinus Cyclostomata; Hyperoartia; Petromyzontiformes; Petromyzontidae; Petromyzon Petromyzon_marinus.0.pep1 Callorhinchus_milii Gnathostomata; Chondrichthyes; Holocephali; Chimaeriformes; Callorhinchidae; Callorhinchus Callorhinchus_milii.0.pep1 Callorhinchus_milii Gnathostomata; Chondrichthyes; Holocephali; Chimaeriformes; Callorhinchidae; Callorhinchus Callorhinchus_milii.0.pep2 Callorhinchus_milii Gnathostomata; Chondrichthyes; Holocephali; Chimaeriformes; Callorhinchidae; Callorhinchus Callorhinchus_milii.0.pep3 Lepisosteus_oculatus Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Holostei; Semionotiformes; Lepisosteus_oculatus.0.pep1 Lepisosteus_oculatus Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Holostei; Semionotiformes; Lepisosteus_oculatus.0.pep2 Lepisosteus_oculatus Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Holostei; Semionotiformes; Lepisosteus_oculatus.0.pep3 Lepisosteus_oculatus Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Holostei; Semionotiformes; Lepisosteus_oculatus.1.pep1 TILa Cynoglossus_semilaevis Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Cynoglossus_semilaevis.1.pep1 -

Transcriptome Ortholog Alignment Sequence Tools (TOAST) for Phylogenomic Dataset Assembly
Transcriptome Ortholog Alignment Sequence Tools (TOAST) for Phylogenomic Dataset Assembly Dustin J. Wcisel North Carolina State University J. Thomas Howard North Carolina State University Jeffrey A. Yoder North Carolina State University Alex Dornburg ( [email protected] ) NC Museum of Natural Sciences https://orcid.org/0000-0003-0863-2283 Software Keywords: BUSCO ortholog assembly, Cetacean and teleost sh phylogeny, Missing Data Visualization, Transcriptome, Concatenated Alignment Posted Date: March 12th, 2020 DOI: https://doi.org/10.21203/rs.2.16269/v4 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Version of Record: A version of this preprint was published at BMC Evolutionary Biology on March 30th, 2020. See the published version at https://doi.org/10.1186/s12862-020-01603-w. Page 1/18 Abstract Background Advances in next-generation sequencing technologies have reduced the cost of whole transcriptome analyses, allowing characterization of non-model species at unprecedented levels. The rapid pace of transcriptomic sequencing has driven the public accumulation of a wealth of data for phylogenomic analyses, however lack of tools aimed towards phylogeneticists to eciently identify orthologous sequences currently hinders effective harnessing of this resource. Results We introduce TOAST, an open source R software package that can utilize the ortholog searches based on the software Benchmarking Universal Single-Copy Orthologs (BUSCO) to assemble multiple sequence alignments of orthologous loci from transcriptomes for any group of organisms. By streamlining search, query, and alignment, TOAST automates the generation of locus and concatenated alignments, and also presents a series of outputs from which users can not only explore missing data patterns across their alignments, but also reassemble alignments based on user-dened acceptable missing data levels for a given research question. -

Origin, Evolution and Homologies of the Weberian Apparatus: a New Insight
Int. J. Morphol., 27(2):333-354, 2009. Origin, Evolution and Homologies of the Weberian Apparatus: A New Insight Origen, Evolución y Homologías del Aparato Weberiano: Un Nuevo Acercamiento Rui Diogo DIOGO, R. Origin, evolution and homologies of the Weberian apparatus: a new insight. Int. J. Morphol., 27(2):333-354, 2009. SUMMARY: The Weberian apparatus is essentially a mechanical device improving audition, consisting of a double chain of ossicles joining the air bladder to the inner ear. Despite being one of the most notable complex systems of teleost fishes and the subject of several comparative, developmental and functional studies, there is still much controversy concerning the origin, evolution and homologies of the structures forming this apparatus. In this paper I provide a new insight on these topics, which takes into account the results of recent works on comparative anatomy, paleontology, and ontogeny as well as of a recent extensive phylogenetic analysis including not only numerous otophysan and non-otophysan extant otocephalans but also ostariophysan fossils such as †Chanoides macropoma, †Clupavus maroccanus, †Santanichthys diasii, †Lusitanichthys characiformis, †Sorbininardus apuliensis and †Tischlingerichthys viohli. According to the evidence now available, the Weberian apparatus of otophysans seems to be the outcome of a functional integration of features acquired in basal otocephalans and in basal ostariophysans, which were very likely not directly related with the functioning of this apparatus, and of features acquired in the nodes leading to the Otophysi and to the clade including the four extant otophysan orders, which could well have been the result of a selection directly related to the functioning of the apparatus. -

Constraints on the Timescale of Animal Evolutionary History
Palaeontologia Electronica palaeo-electronica.org Constraints on the timescale of animal evolutionary history Michael J. Benton, Philip C.J. Donoghue, Robert J. Asher, Matt Friedman, Thomas J. Near, and Jakob Vinther ABSTRACT Dating the tree of life is a core endeavor in evolutionary biology. Rates of evolution are fundamental to nearly every evolutionary model and process. Rates need dates. There is much debate on the most appropriate and reasonable ways in which to date the tree of life, and recent work has highlighted some confusions and complexities that can be avoided. Whether phylogenetic trees are dated after they have been estab- lished, or as part of the process of tree finding, practitioners need to know which cali- brations to use. We emphasize the importance of identifying crown (not stem) fossils, levels of confidence in their attribution to the crown, current chronostratigraphic preci- sion, the primacy of the host geological formation and asymmetric confidence intervals. Here we present calibrations for 88 key nodes across the phylogeny of animals, rang- ing from the root of Metazoa to the last common ancestor of Homo sapiens. Close attention to detail is constantly required: for example, the classic bird-mammal date (base of crown Amniota) has often been given as 310-315 Ma; the 2014 international time scale indicates a minimum age of 318 Ma. Michael J. Benton. School of Earth Sciences, University of Bristol, Bristol, BS8 1RJ, U.K. [email protected] Philip C.J. Donoghue. School of Earth Sciences, University of Bristol, Bristol, BS8 1RJ, U.K. [email protected] Robert J. -
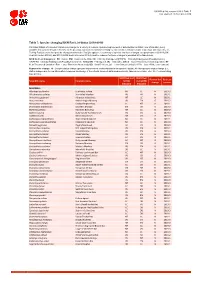
Table 7: Species Changing IUCN Red List Status (2018-2019)
IUCN Red List version 2019-3: Table 7 Last Updated: 10 December 2019 Table 7: Species changing IUCN Red List Status (2018-2019) Published listings of a species' status may change for a variety of reasons (genuine improvement or deterioration in status; new information being available that was not known at the time of the previous assessment; taxonomic changes; corrections to mistakes made in previous assessments, etc. To help Red List users interpret the changes between the Red List updates, a summary of species that have changed category between 2018 (IUCN Red List version 2018-2) and 2019 (IUCN Red List version 2019-3) and the reasons for these changes is provided in the table below. IUCN Red List Categories: EX - Extinct, EW - Extinct in the Wild, CR - Critically Endangered [CR(PE) - Critically Endangered (Possibly Extinct), CR(PEW) - Critically Endangered (Possibly Extinct in the Wild)], EN - Endangered, VU - Vulnerable, LR/cd - Lower Risk/conservation dependent, NT - Near Threatened (includes LR/nt - Lower Risk/near threatened), DD - Data Deficient, LC - Least Concern (includes LR/lc - Lower Risk, least concern). Reasons for change: G - Genuine status change (genuine improvement or deterioration in the species' status); N - Non-genuine status change (i.e., status changes due to new information, improved knowledge of the criteria, incorrect data used previously, taxonomic revision, etc.); E - Previous listing was an Error. IUCN Red List IUCN Red Reason for Red List Scientific name Common name (2018) List (2019) change version Category -

Percomorph Phylogeny: a Survey of Acanthomorphs and a New Proposal
BULLETIN OF MARINE SCIENCE, 52(1): 554-626, 1993 PERCOMORPH PHYLOGENY: A SURVEY OF ACANTHOMORPHS AND A NEW PROPOSAL G. David Johnson and Colin Patterson ABSTRACT The interrelationships of acanthomorph fishes are reviewed. We recognize seven mono- phyletic terminal taxa among acanthomorphs: Lampridiformes, Polymixiiformes, Paracan- thopterygii, Stephanoberyciformes, Beryciformes, Zeiformes, and a new taxon named Smeg- mamorpha. The Percomorpha, as currently constituted, are polyphyletic, and the Perciformes are probably paraphyletic. The smegmamorphs comprise five subgroups: Synbranchiformes (Synbranchoidei and Mastacembeloidei), Mugilomorpha (Mugiloidei), Elassomatidae (Elas- soma), Gasterosteiformes, and Atherinomorpha. Monophyly of Lampridiformes is justified elsewhere; we have found no new characters to substantiate the monophyly of Polymixi- iformes (which is not in doubt) or Paracanthopterygii. Stephanoberyciformes uniquely share a modification of the extrascapular, and Beryciformes a modification of the anterior part of the supraorbital and infraorbital sensory canals, here named Jakubowski's organ. Our Zei- formes excludes the Caproidae, and characters are proposed to justify the monophyly of the group in that restricted sense. The Smegmamorpha are thought to be monophyletic principally because of the configuration of the first vertebra and its intermuscular bone. Within the Smegmamorpha, the Atherinomorpha and Mugilomorpha are shown to be monophyletic elsewhere. Our Gasterosteiformes includes the syngnathoids and the Pegasiformes