The Spliceosome-Activating Complex: Molecular Mechanisms Underlying the Function of a Pleiotropic Regulator
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

PLRG1 Antibody Cat
PLRG1 Antibody Cat. No.: 46-203 PLRG1 Antibody 46-203 (5ug/ml) staining of paraffin embedded Human Prostate. Steamed antigen retrieval with citrate buffer pH 6, AP-staining. Specifications HOST SPECIES: Goat SPECIES REACTIVITY: Human, Mouse HOMOLOGY: Expected Species Reactivity based on sequence homology: Rat, Dog, Pig, Cow IMMUNOGEN: The immunogen for this antibody is: C-PVSWKPEIIKRKRF TESTED APPLICATIONS: ELISA, IHC, WB Peptide ELISA: antibody detection limit dilution 1:16000.Western Blot:Approx 60kDa band observed in nuclear lysates of cell lines HEK293 and NIH3T3 (calculated MW of 57.2kDa according to Human NP_002660.1 and 56.9kDa according to Mouse APPLICATIONS: NP_058064.2). Recommended concentration: 0.1-0.3ug/ml. Primary incubation was 1 hour.Immunohistochemistry:Paraffin embedded Human Prostate. Recommended concentration: 5ug/ml. September 28, 2021 1 https://www.prosci-inc.com/plrg1-antibody-46-203.html This antibody is expected to recognise isoform 1 (NP_002660.1) and isoform 2 SPECIFICITY: (NP_001188493.1). POSITIVE CONTROL: 1) Cat. No. 1405 - Mouse Kidney Tissue Lysate Properties Purified from goat serum by ammonium sulphate precipitation followed by antigen PURIFICATION: affinity chromatography using the immunizing peptide. CLONALITY: Polyclonal CONJUGATE: Unconjugated PHYSICAL STATE: Liquid Supplied at 0.5 mg/ml in Tris saline, 0.02% sodium azide, pH7.3 with 0.5% bovine serum BUFFER: albumin. Aliquot and store at -20°C. Minimize freezing and thawing. CONCENTRATION: 500 ug/mL STORAGE CONDITIONS: Aliquot and store at -20˚C. -
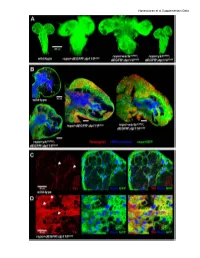
Supplementary Data Vigneswaran Et Al Supplementary Data
Vigneswaran et al Supplementary Data Vigneswaran et al Supplementary Data Figure S1: Yki is required for EGFR-PI3K-driven glial neoplasia in Drosophila (A) Optical projections of whole brain-nerve cord complexes from 3rd instar larvae approximately 130 hrs old. Dorsal view; anterior up. CD8-GFP (green) labels glial cell bodies. Compared to repo>dEGFRλ;dp110CAAX, warts knockdown (repo>wartsdsRNA; dEGFRλ;dp110CAAX) increased neoplastic brain overgrowth and yki knockdown (repo>ykidsRNA;dEGFRλ;dp110CAAX) decreased neoplastic brain overgrowth. (B) 3 µm optical projections of brain hemispheres, age-matched 3rd instar larvae. Frontal sections; anterior up; midline to left. Repo (red) labels glial cell nuclei; CD8-GFP (green) labels glial cell bodies; anti-HRP (blue) counter-stains for neurons and neuropil. (middle) repo>dEGFRλ;dp110CAAX showed increased glial cell numbers (red nuclei) compared to (upper left) wild-type. Compared to repo>dEGFRλ;dp110CAAX, (right) warts knockdown increased neoplastic glial cell numbers (red nuclei), whereas (lower left) yki knockdown reduced neoplastic glial cell numbers (red nuclei). (C, D) Low levels of Yki protein (red) was observed in wild-type central brain glia (white arrows, left panel in C) compared to high levels of cytoplasmic and nuclear Yki protein in dEGFRλ;dp110CAAX neoplastic glia (white arrows, left panel in D); Repo (blue) labels glial cell nuclei; CD8-GFP (green) labels glial cell bodies. Vigneswaran et al Supplementary Data Figure S2: YAP/TAZ expression confined to RTK-amplified tuMor cells and Maintained in patient-derived xenografts (A) On the left, immunohistochemical (IHC) staining in representative normal brain parenchyma in the cortex where YAP expression and TAZ expression was limited to vascular cells and was not detectable in normal neuronal and glial cells. -

Direct Interaction Between Hnrnp-M and CDC5L/PLRG1 Proteins Affects Alternative Splice Site Choice
Direct interaction between hnRNP-M and CDC5L/PLRG1 proteins affects alternative splice site choice David Llères, Marco Denegri, Marco Biggiogera, Paul Ajuh, Angus Lamond To cite this version: David Llères, Marco Denegri, Marco Biggiogera, Paul Ajuh, Angus Lamond. Direct interaction be- tween hnRNP-M and CDC5L/PLRG1 proteins affects alternative splice site choice. EMBO Reports, EMBO Press, 2010, 11 (6), pp.445 - 451. 10.1038/embor.2010.64. hal-03027049 HAL Id: hal-03027049 https://hal.archives-ouvertes.fr/hal-03027049 Submitted on 26 Nov 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. scientificscientificreport report Direct interaction between hnRNP-M and CDC5L/ PLRG1 proteins affects alternative splice site choice David Lle`res1*, Marco Denegri1*w,MarcoBiggiogera2,PaulAjuh1z & Angus I. Lamond1+ 1Wellcome Trust Centre for Gene Regulation & Expression, College of Life Sciences, University of Dundee, Dundee, UK, and 2LaboratoriodiBiologiaCellulareandCentrodiStudioperl’IstochimicadelCNR,DipartimentodiBiologiaAnimale, Universita’ di Pavia, Pavia, Italy Heterogeneous nuclear ribonucleoprotein-M (hnRNP-M) is an and affect the fate of heterogeneous nuclear RNAs by influencing their abundant nuclear protein that binds to pre-mRNA and is a structure and/or by facilitating or hindering the interaction of their component of the spliceosome complex. -

Mrna Editing, Processing and Quality Control in Caenorhabditis Elegans
| WORMBOOK mRNA Editing, Processing and Quality Control in Caenorhabditis elegans Joshua A. Arribere,*,1 Hidehito Kuroyanagi,†,1 and Heather A. Hundley‡,1 *Department of MCD Biology, UC Santa Cruz, California 95064, †Laboratory of Gene Expression, Medical Research Institute, Tokyo Medical and Dental University, Tokyo 113-8510, Japan, and ‡Medical Sciences Program, Indiana University School of Medicine-Bloomington, Indiana 47405 ABSTRACT While DNA serves as the blueprint of life, the distinct functions of each cell are determined by the dynamic expression of genes from the static genome. The amount and specific sequences of RNAs expressed in a given cell involves a number of regulated processes including RNA synthesis (transcription), processing, splicing, modification, polyadenylation, stability, translation, and degradation. As errors during mRNA production can create gene products that are deleterious to the organism, quality control mechanisms exist to survey and remove errors in mRNA expression and processing. Here, we will provide an overview of mRNA processing and quality control mechanisms that occur in Caenorhabditis elegans, with a focus on those that occur on protein-coding genes after transcription initiation. In addition, we will describe the genetic and technical approaches that have allowed studies in C. elegans to reveal important mechanistic insight into these processes. KEYWORDS Caenorhabditis elegans; splicing; RNA editing; RNA modification; polyadenylation; quality control; WormBook TABLE OF CONTENTS Abstract 531 RNA Editing and Modification 533 Adenosine-to-inosine RNA editing 533 The C. elegans A-to-I editing machinery 534 RNA editing in space and time 535 ADARs regulate the levels and fates of endogenous dsRNA 537 Are other modifications present in C. -

Structural Insights Into Nuclear Pre-Mrna Splicing in Higher Eukaryotes
Downloaded from http://cshperspectives.cshlp.org/ on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Structural Insights into Nuclear pre-mRNA Splicing in Higher Eukaryotes Berthold Kastner,1 Cindy L. Will,1 Holger Stark,2 and Reinhard Lührmann1 1Department of Cellular Biochemistry, Max Planck Institute for Biophysical Chemistry, D-37077 Göttingen, Germany 2Department of Structural Dynamics, Max Planck Institute for Biophysical Chemistry, D-37077 Göttingen, Germany Correspondence: [email protected] SUMMARY The spliceosome is a highly complex, dynamic ribonucleoprotein molecular machine that undergoes numerous structural and compositional rearrangements that lead to the formation of its active site. Recent advances in cyroelectron microscopy (cryo-EM) have provided a plethora of near-atomic structural information about the inner workings of the spliceosome. Aided by previous biochemical, structural, and functional studies, cryo-EM has confirmed or provided a structural basis for most of the prevailing models of spliceosome function, but at the same time allowed novel insights into splicing catalysis and the intriguing dynamics of the spliceosome. The mechanism of pre-mRNA splicing is highly conserved between humans and yeast, but the compositional dynamics and ribonucleoprotein (RNP) remodeling of the human spliceosome are more complex. Here, we summarize recent advances in our understanding of the molec- ular architecture of the human spliceosome, highlighting differences between the human and yeast -

Downregulation of Carnitine Acyl-Carnitine Translocase by Mirnas
Page 1 of 288 Diabetes 1 Downregulation of Carnitine acyl-carnitine translocase by miRNAs 132 and 212 amplifies glucose-stimulated insulin secretion Mufaddal S. Soni1, Mary E. Rabaglia1, Sushant Bhatnagar1, Jin Shang2, Olga Ilkayeva3, Randall Mynatt4, Yun-Ping Zhou2, Eric E. Schadt6, Nancy A.Thornberry2, Deborah M. Muoio5, Mark P. Keller1 and Alan D. Attie1 From the 1Department of Biochemistry, University of Wisconsin, Madison, Wisconsin; 2Department of Metabolic Disorders-Diabetes, Merck Research Laboratories, Rahway, New Jersey; 3Sarah W. Stedman Nutrition and Metabolism Center, Duke Institute of Molecular Physiology, 5Departments of Medicine and Pharmacology and Cancer Biology, Durham, North Carolina. 4Pennington Biomedical Research Center, Louisiana State University system, Baton Rouge, Louisiana; 6Institute for Genomics and Multiscale Biology, Mount Sinai School of Medicine, New York, New York. Corresponding author Alan D. Attie, 543A Biochemistry Addition, 433 Babcock Drive, Department of Biochemistry, University of Wisconsin-Madison, Madison, Wisconsin, (608) 262-1372 (Ph), (608) 263-9608 (fax), [email protected]. Running Title: Fatty acyl-carnitines enhance insulin secretion Abstract word count: 163 Main text Word count: 3960 Number of tables: 0 Number of figures: 5 Diabetes Publish Ahead of Print, published online June 26, 2014 Diabetes Page 2 of 288 2 ABSTRACT We previously demonstrated that micro-RNAs 132 and 212 are differentially upregulated in response to obesity in two mouse strains that differ in their susceptibility to obesity-induced diabetes. Here we show the overexpression of micro-RNAs 132 and 212 enhances insulin secretion (IS) in response to glucose and other secretagogues including non-fuel stimuli. We determined that carnitine acyl-carnitine translocase (CACT, Slc25a20) is a direct target of these miRNAs. -

Alternatively Spliced Genes
1 Alternatively Spliced Genes Jane Y. Wu1,2,LiyaYuan1 and Necat Havlioglu1 1Washington University School of Medicine, St. Louis, MO, USA 2John F. Kennedy Center for Research on Human Development, Vanderbilt University Medical Center, Nashville, TN, USA 1 Pre-mRNA Splicing and Splicing Machinery 3 1.1 Splicing Machinery: Spliceosome 3 1.2 Splicing Signals 4 1.3 Spliceosomal UsnRNP Biogenesis 12 1.4 Spliceosome Assembly 13 1.5 Biochemical Mechanisms of pre-mRNA Splicing 17 2 Alternative pre-mRNA Splicing 17 2.1 Alternative Splicing and its Role in Regulating Gene Activities and Generating Genetic Diversity 17 2.1.1 Different Patterns of Alternative Splicing 17 2.1.2 Alternative Splicing and Genetic Diversity 18 2.2 Mechanisms Underlying Alternative Splicing Regulation 19 2.2.1 Splicing Signals and Splicing Regulatory Elements 20 2.2.2 Trans-acting Splicing Regulators 23 2.3 Tissue-specific and Developmentally Regulated Alternative Splicing 26 2.4 Regulation of Alternative Splicing in Response to Extracellular Stimuli 27 3 Pre-mRNA Splicing and Human Diseases 28 3.1 Splicing Defects in Human Diseases 28 3.2 Molecular Mechanisms Underlying Splicing Defects Associated with Disease 33 4 Perspectives on Diagnosis and Treatment of Diseases Caused by pre-mRNA Splicing Defects 36 4.1 Diagnosis of Human Diseases Caused by Splicing Defects 36 2 Alternatively Spliced Genes 4.2 Potential Therapeutic Approaches 37 4.2.1 Oligonucleotide-based Approaches: Antisense, RNAi, and Chimeric Molecules 37 4.2.2 Ribozymes 37 4.2.3 SMaRT 38 4.2.4 Chemical Compounds 38 5 Concluding Remarks 38 Acknowledgment 39 Bibliography 39 Books and Reviews 39 Keywords Pre-mRNA Nascent transcripts that are precursors of mature messenger RNAs. -

CTNNBL1 Facilitates the Association of CWC15 with CDC5L and Is
7058–7069 Nucleic Acids Research, 2015, Vol. 43, No. 14 Published online 29 June 2015 doi: 10.1093/nar/gkv643 CTNNBL1 facilitates the association of CWC15 with CDC5L and is required to maintain the abundance of the Prp19 spliceosomal complex Febe van Maldegem1,*, Sarah Maslen1, Christopher M. Johnson1, Anita Chandra1, Karuna Ganesh2, Mark Skehel1 and Cristina Rada1,* 1MRC Laboratory of Molecular Biology, Cambridge, CB2 0QH, UK and 2Department of Medicine and Cancer Biology & Genetics Program, Memorial Sloan Kettering Cancer Center, New York, NY 10065, USA Received November 18, 2014; Revised June 04, 2015; Accepted June 09, 2015 ABSTRACT CDC5L, PLRG1 and SPF27 (the product of the BCAS2 gene), and a sub-complex that includes CTNNBL1 and In order to catalyse the splicing of messenger RNA, CWC15, as well as the chaperone protein HSP7C (en- multiple proteins and RNA components associate coded by the HSPA8 gene) (2). As part of a bigger com- and dissociate in a dynamic highly choreographed plex of more than 30 associated proteins (3), the Prp19 process. The Prp19 complex is a conserved essential complex aids the progression of the core snRNP compo- part of the splicing machinery thought to facilitate the nents through several steps of catalysis and recycling and conformational changes the spliceosome undergoes is therefore essential for splicing (4). The core complex during catalysis. Dynamic protein interactions often has remarkable conservation of function, being essential involve highly disordered regions that are difficult to in both Saccharomyces cerevisiae and Schizosaccharomyces study by structural methods. Using amine crosslink- pombe––known as the NineTeen Complex or NTC (5). -

Biology of the Mrna Splicing Machinery and Its Dysregulation in Cancer Providing Therapeutic Opportunities
International Journal of Molecular Sciences Review Biology of the mRNA Splicing Machinery and Its Dysregulation in Cancer Providing Therapeutic Opportunities Maxime Blijlevens †, Jing Li † and Victor W. van Beusechem * Medical Oncology, Amsterdam UMC, Cancer Center Amsterdam, Vrije Universiteit Amsterdam, de Boelelaan 1117, 1081 HV Amsterdam, The Netherlands; [email protected] (M.B.); [email protected] (J.L.) * Correspondence: [email protected]; Tel.: +31-2044-421-62 † Shared first author. Abstract: Dysregulation of messenger RNA (mRNA) processing—in particular mRNA splicing—is a hallmark of cancer. Compared to normal cells, cancer cells frequently present aberrant mRNA splicing, which promotes cancer progression and treatment resistance. This hallmark provides opportunities for developing new targeted cancer treatments. Splicing of precursor mRNA into mature mRNA is executed by a dynamic complex of proteins and small RNAs called the spliceosome. Spliceosomes are part of the supraspliceosome, a macromolecular structure where all co-transcriptional mRNA processing activities in the cell nucleus are coordinated. Here we review the biology of the mRNA splicing machinery in the context of other mRNA processing activities in the supraspliceosome and present current knowledge of its dysregulation in lung cancer. In addition, we review investigations to discover therapeutic targets in the spliceosome and give an overview of inhibitors and modulators of the mRNA splicing process identified so far. Together, this provides insight into the value of targeting the spliceosome as a possible new treatment for lung cancer. Citation: Blijlevens, M.; Li, J.; van Beusechem, V.W. Biology of the Keywords: alternative splicing; splicing dysregulation; splicing factors; NSCLC mRNA Splicing Machinery and Its Dysregulation in Cancer Providing Therapeutic Opportunities. -

Protein List
Protein Accession Protein Id Protein Name P11171 41 Protein 4. -

“Characterisation of the Biflavonoid Hinokiflavone As a Pre-Mrna
bioRxiv preprint doi: https://doi.org/10.1101/123026; this version posted April 1, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. “Characterisation of the biflavonoid hinokiflavone as a pre- mRNA splicing modulator that inhibits SENP” Andrea Pawellek1, Ursula Ryder1, Triin Tammsalu1, Lewis J. King2 Helmi Kreinin2, Tony Ly1, Ronald T. Hay1, Richard Hartley2 and Angus I. Lamond1 1University of Dundee, School of Life Sciences, Centre for Gene Regulation and Expression, Dundee, DD1 5EH, UK, 2University of Glasgow, WestCHEM School of Chemistry, Glasgow, G12 8QQ, UK 1 bioRxiv preprint doi: https://doi.org/10.1101/123026; this version posted April 1, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Here, we identify the plant biflavanoid hinokiflavone as an inhibitor of splicing in vitro and modulater of alternative splicing in multiple human cell lines. Hinokiflavone inhibits splicing in vitro by blocking one or more early steps of spliceosome assembly, leading to accumulation of the A complex. Multiple human cell lines treated with hinokiflavone show changes in the alternative splicing of different pre-mRNA substrates, but little or no change in transcription. They also show altered subnuclear organization, specifically of splicing factors required for A complex formation, which relocalized together with SUMO1 and SUMO2 into enlarged nuclear speckles. While most cell lines treated with hinokiflavone showed cell cycle arrest and eventual cell death, dependent on time and concentration, the promyelocytic NB4 cell line, which expresses the SUMO target PML-RARalpha fusion protein, was exquisitely sensitive to apoptosis following hinokiflavone treatment.