“Characterisation of the Biflavonoid Hinokiflavone As a Pre-Mrna
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Premature Termination Codons in PRPF31 Cause Retinitis Pigmentosa Via Haploinsufficiency Due to Nonsense-Mediated Mrna Decay
Premature termination codons in PRPF31 cause retinitis pigmentosa via haploinsufficiency due to nonsense-mediated mRNA decay Thomas Rio Frio, … , Jacques S. Beckmann, Carlo Rivolta J Clin Invest. 2008;118(4):1519-1531. https://doi.org/10.1172/JCI34211. Research Article Genetics Dominant mutations in the gene encoding the mRNA splicing factor PRPF31 cause retinitis pigmentosa, a hereditary form of retinal degeneration. Most of these mutations are characterized by DNA changes that lead to premature termination codons. We investigated 6 different PRPF31 mutations, represented by single-base substitutions or microdeletions, in cell lines derived from 9 patients with dominant retinitis pigmentosa. Five of these mutations lead to premature termination codons, and 1 leads to the skipping of exon 2. Allele-specific measurement of PRPF31 transcripts revealed a strong reduction in the expression of mutant alleles. As a consequence, total PRPF31 protein abundance was decreased, and no truncated proteins were detected. Subnuclear localization of the full-length PRPF31 that was present remained unaffected. Blocking nonsense-mediated mRNA decay significantly restored the amount of mutant PRPF31 mRNA but did not restore the synthesis of mutant proteins, even in conjunction with inhibitors of protein degradation pathways. Our results indicate that most PRPF31 mutations ultimately result in null alleles through the activation of surveillance mechanisms that inactivate mutant mRNA and, possibly, proteins. Furthermore, these data provide compelling evidence that the pathogenic effect of PRPF31 mutations is likely due to haploinsufficiency rather than to gain of function. Find the latest version: https://jci.me/34211/pdf Research article Premature termination codons in PRPF31 cause retinitis pigmentosa via haploinsufficiency due to nonsense-mediated mRNA decay Thomas Rio Frio,1 Nicholas M. -

Miasdb: a Database of Molecular Interactions Associated with Alternative Splicing of Human Pre-Mrnas
RESEARCH ARTICLE MiasDB: A Database of Molecular Interactions Associated with Alternative Splicing of Human Pre-mRNAs Yongqiang Xing1, Xiujuan Zhao1, Tao Yu2, Dong Liang1, Jun Li1, Guanyun Wei1, Guoqing Liu1, Xiangjun Cui1, Hongyu Zhao1, Lu Cai1* 1 School of Life Science and Technology, Inner Mongolia University of Science and Technology, Baotou, 014010, China, 2 School of Science, Inner Mongolia University of Science and Technology, Baotou, 014010, China a11111 * [email protected] Abstract Alternative splicing (AS) is pervasive in human multi-exon genes and is a major contributor to expansion of the transcriptome and proteome diversity. The accurate recognition of alter- OPEN ACCESS native splice sites is regulated by information contained in networks of protein-protein and Citation: Xing Y, Zhao X, Yu T, Liang D, Li J, Wei G, protein-RNA interactions. However, the mechanisms leading to splice site selection are not et al. (2016) MiasDB: A Database of Molecular fully understood. Although numerous databases have been built to describe AS, molecular Interactions Associated with Alternative Splicing of Human Pre-mRNAs. PLoS ONE 11(5): e0155443. interaction databases associated with AS have only recently emerged. In this study, we doi:10.1371/journal.pone.0155443 present a new database, MiasDB, that provides a description of molecular interactions Editor: Ruben Artero, University of Valencia, SPAIN associated with human AS events. This database covers 938 interactions between human splicing factors, RNA elements, transcription factors, kinases and modified histones for 173 Received: November 19, 2015 human AS events. Every entry includes the interaction partners, interaction type, experi- Accepted: April 28, 2016 mental methods, AS type, tissue specificity or disease-relevant information, a simple Published: May 11, 2016 description of the functionally tested interaction in the AS event and references. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Transcriptome Sequencing and Genome-Wide Association Analyses Reveal Lysosomal Function and Actin Cytoskeleton Remodeling in Schizophrenia and Bipolar Disorder
Molecular Psychiatry (2015) 20, 563–572 © 2015 Macmillan Publishers Limited All rights reserved 1359-4184/15 www.nature.com/mp ORIGINAL ARTICLE Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder Z Zhao1,6,JXu2,6, J Chen3,6, S Kim4, M Reimers3, S-A Bacanu3,HYu1, C Liu5, J Sun1, Q Wang1, P Jia1,FXu2, Y Zhang2, KS Kendler3, Z Peng2 and X Chen3 Schizophrenia (SCZ) and bipolar disorder (BPD) are severe mental disorders with high heritability. Clinicians have long noticed the similarities of clinic symptoms between these disorders. In recent years, accumulating evidence indicates some shared genetic liabilities. However, what is shared remains elusive. In this study, we conducted whole transcriptome analysis of post-mortem brain tissues (cingulate cortex) from SCZ, BPD and control subjects, and identified differentially expressed genes in these disorders. We found 105 and 153 genes differentially expressed in SCZ and BPD, respectively. By comparing the t-test scores, we found that many of the genes differentially expressed in SCZ and BPD are concordant in their expression level (q ⩽ 0.01, 53 genes; q ⩽ 0.05, 213 genes; q ⩽ 0.1, 885 genes). Using genome-wide association data from the Psychiatric Genomics Consortium, we found that these differentially and concordantly expressed genes were enriched in association signals for both SCZ (Po10 − 7) and BPD (P = 0.029). To our knowledge, this is the first time that a substantially large number of genes show concordant expression and association for both SCZ and BPD. Pathway analyses of these genes indicated that they are involved in the lysosome, Fc gamma receptor-mediated phagocytosis, regulation of actin cytoskeleton pathways, along with several cancer pathways. -

Genes with 5' Terminal Oligopyrimidine Tracts Preferentially Escape Global Suppression of Translation by the SARS-Cov-2 NSP1 Protein
Downloaded from rnajournal.cshlp.org on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Genes with 5′ terminal oligopyrimidine tracts preferentially escape global suppression of translation by the SARS-CoV-2 Nsp1 protein Shilpa Raoa, Ian Hoskinsa, Tori Tonna, P. Daniela Garciaa, Hakan Ozadama, Elif Sarinay Cenika, Can Cenika,1 a Department of Molecular Biosciences, University of Texas at Austin, Austin, TX 78712, USA 1Corresponding author: [email protected] Key words: SARS-CoV-2, Nsp1, MeTAFlow, translation, ribosome profiling, RNA-Seq, 5′ TOP, Ribo-Seq, gene expression 1 Downloaded from rnajournal.cshlp.org on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Abstract Viruses rely on the host translation machinery to synthesize their own proteins. Consequently, they have evolved varied mechanisms to co-opt host translation for their survival. SARS-CoV-2 relies on a non-structural protein, Nsp1, for shutting down host translation. However, it is currently unknown how viral proteins and host factors critical for viral replication can escape a global shutdown of host translation. Here, using a novel FACS-based assay called MeTAFlow, we report a dose-dependent reduction in both nascent protein synthesis and mRNA abundance in cells expressing Nsp1. We perform RNA-Seq and matched ribosome profiling experiments to identify gene-specific changes both at the mRNA expression and translation level. We discover that a functionally-coherent subset of human genes are preferentially translated in the context of Nsp1 expression. These genes include the translation machinery components, RNA binding proteins, and others important for viral pathogenicity. Importantly, we uncovered a remarkable enrichment of 5′ terminal oligo-pyrimidine (TOP) tracts among preferentially translated genes. -

1 AGING Supplementary Table 2
SUPPLEMENTARY TABLES Supplementary Table 1. Details of the eight domain chains of KIAA0101. Serial IDENTITY MAX IN COMP- INTERFACE ID POSITION RESOLUTION EXPERIMENT TYPE number START STOP SCORE IDENTITY LEX WITH CAVITY A 4D2G_D 52 - 69 52 69 100 100 2.65 Å PCNA X-RAY DIFFRACTION √ B 4D2G_E 52 - 69 52 69 100 100 2.65 Å PCNA X-RAY DIFFRACTION √ C 6EHT_D 52 - 71 52 71 100 100 3.2Å PCNA X-RAY DIFFRACTION √ D 6EHT_E 52 - 71 52 71 100 100 3.2Å PCNA X-RAY DIFFRACTION √ E 6GWS_D 41-72 41 72 100 100 3.2Å PCNA X-RAY DIFFRACTION √ F 6GWS_E 41-72 41 72 100 100 2.9Å PCNA X-RAY DIFFRACTION √ G 6GWS_F 41-72 41 72 100 100 2.9Å PCNA X-RAY DIFFRACTION √ H 6IIW_B 2-11 2 11 100 100 1.699Å UHRF1 X-RAY DIFFRACTION √ www.aging-us.com 1 AGING Supplementary Table 2. Significantly enriched gene ontology (GO) annotations (cellular components) of KIAA0101 in lung adenocarcinoma (LinkedOmics). Leading Description FDR Leading Edge Gene EdgeNum RAD51, SPC25, CCNB1, BIRC5, NCAPG, ZWINT, MAD2L1, SKA3, NUF2, BUB1B, CENPA, SKA1, AURKB, NEK2, CENPW, HJURP, NDC80, CDCA5, NCAPH, BUB1, ZWILCH, CENPK, KIF2C, AURKA, CENPN, TOP2A, CENPM, PLK1, ERCC6L, CDT1, CHEK1, SPAG5, CENPH, condensed 66 0 SPC24, NUP37, BLM, CENPE, BUB3, CDK2, FANCD2, CENPO, CENPF, BRCA1, DSN1, chromosome MKI67, NCAPG2, H2AFX, HMGB2, SUV39H1, CBX3, TUBG1, KNTC1, PPP1CC, SMC2, BANF1, NCAPD2, SKA2, NUP107, BRCA2, NUP85, ITGB3BP, SYCE2, TOPBP1, DMC1, SMC4, INCENP. RAD51, OIP5, CDK1, SPC25, CCNB1, BIRC5, NCAPG, ZWINT, MAD2L1, SKA3, NUF2, BUB1B, CENPA, SKA1, AURKB, NEK2, ESCO2, CENPW, HJURP, TTK, NDC80, CDCA5, BUB1, ZWILCH, CENPK, KIF2C, AURKA, DSCC1, CENPN, CDCA8, CENPM, PLK1, MCM6, ERCC6L, CDT1, HELLS, CHEK1, SPAG5, CENPH, PCNA, SPC24, CENPI, NUP37, FEN1, chromosomal 94 0 CENPL, BLM, KIF18A, CENPE, MCM4, BUB3, SUV39H2, MCM2, CDK2, PIF1, DNA2, region CENPO, CENPF, CHEK2, DSN1, H2AFX, MCM7, SUV39H1, MTBP, CBX3, RECQL4, KNTC1, PPP1CC, CENPP, CENPQ, PTGES3, NCAPD2, DYNLL1, SKA2, HAT1, NUP107, MCM5, MCM3, MSH2, BRCA2, NUP85, SSB, ITGB3BP, DMC1, INCENP, THOC3, XPO1, APEX1, XRCC5, KIF22, DCLRE1A, SEH1L, XRCC3, NSMCE2, RAD21. -

PRPF40A As a Potential Diagnostic and Prognostic Marker Is Upregulated in Pancreatic Cancer Tissues and Cell Lines: an Integrated Bioinformatics Data Analysis
OncoTargets and Therapy Dovepress open access to scientific and medical research Open Access Full Text Article ORIGINAL RESEARCH PRPF40A as a potential diagnostic and prognostic marker is upregulated in pancreatic cancer tissues and cell lines: an integrated bioinformatics data analysis This article was published in the following Dove Press journal: OncoTargets and Therapy Zhen Huo* Background: Pre-mRNA processing factor 40 homolog A (PRPF40A) is an important Shuyu Zhai* protein involved in pre-mRNA splicing and is expressed in a variety of cell types. Yuanchi Weng* However, the function of PRPF40A in pancreatic cancer remains unclear. Therefore, our Hao Qian study is to investigate the role of PRPF40A in the pathogenesis of pancreatic cancer. Xiaomei Tang Materials and methods: We extracted expression data and clinical information of Yusheng Shi PRPF40A from different online databases, including the Cancer Genome Atlas (TCGA), Oncomine and the Gene Expression Omnibus (GEO). Subsequently, samples were collected Xiaxing Deng from patients to validate gene expression using qPCR, Western blotting and immunohisto- Yue Wang chemical (IHC) analyses. Receiver operating characteristic (ROC) and Kaplan-Meier curve Baiyong Shen were used to evaluate the diagnostic and prognostic potential. Colony formation assays and Department of General Surgery, Ruijin CCK-8 assays were performed to measure the proliferative capacity of pancreatic cancer. Hospital, Shanghai Jiao Tong University Finally, gene ontology (GO) and pathway enrichment analyses of co-expressed genes of School of Medicine, Shanghai 200025, People’s Republic of China PRPF40A were conducted using the Database for Annotation, Visualization and Integrated Discovery (DAVID). *These authors contributed equally to Results: We found that PRPF40A was upregulated based on data from both the online this work databases and our samples. -

WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT (51) International Patent Classification: CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, C12Q 1/68 (2018.01) A61P 31/18 (2006.01) DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, C12Q 1/70 (2006.01) HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, (21) International Application Number: MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, PCT/US2018/056167 OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, (22) International Filing Date: SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, 16 October 2018 (16. 10.2018) TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (26) Publication Language: English GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, TZ, (30) Priority Data: UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, 62/573,025 16 October 2017 (16. 10.2017) US TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, EE, ES, FI, FR, GB, GR, HR, HU, ΓΕ , IS, IT, LT, LU, LV, (71) Applicant: MASSACHUSETTS INSTITUTE OF MC, MK, MT, NL, NO, PL, PT, RO, RS, SE, SI, SK, SM, TECHNOLOGY [US/US]; 77 Massachusetts Avenue, TR), OAPI (BF, BJ, CF, CG, CI, CM, GA, GN, GQ, GW, Cambridge, Massachusetts 02139 (US). -

Nuclear PTEN Safeguards Pre-Mrna Splicing to Link Golgi Apparatus for Its Tumor Suppressive Role
ARTICLE DOI: 10.1038/s41467-018-04760-1 OPEN Nuclear PTEN safeguards pre-mRNA splicing to link Golgi apparatus for its tumor suppressive role Shao-Ming Shen1, Yan Ji2, Cheng Zhang1, Shuang-Shu Dong2, Shuo Yang1, Zhong Xiong1, Meng-Kai Ge1, Yun Yu1, Li Xia1, Meng Guo1, Jin-Ke Cheng3, Jun-Ling Liu1,3, Jian-Xiu Yu1,3 & Guo-Qiang Chen1 Dysregulation of pre-mRNA alternative splicing (AS) is closely associated with cancers. However, the relationships between the AS and classic oncogenes/tumor suppressors are 1234567890():,; largely unknown. Here we show that the deletion of tumor suppressor PTEN alters pre-mRNA splicing in a phosphatase-independent manner, and identify 262 PTEN-regulated AS events in 293T cells by RNA sequencing, which are associated with significant worse outcome of cancer patients. Based on these findings, we report that nuclear PTEN interacts with the splicing machinery, spliceosome, to regulate its assembly and pre-mRNA splicing. We also identify a new exon 2b in GOLGA2 transcript and the exon exclusion contributes to PTEN knockdown-induced tumorigenesis by promoting dramatic Golgi extension and secretion, and PTEN depletion significantly sensitizes cancer cells to secretion inhibitors brefeldin A and golgicide A. Our results suggest that Golgi secretion inhibitors alone or in combination with PI3K/Akt kinase inhibitors may be therapeutically useful for PTEN-deficient cancers. 1 Department of Pathophysiology, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, Shanghai Jiao Tong University School of Medicine (SJTU-SM), Shanghai 200025, China. 2 Institute of Health Sciences, Shanghai Institutes for Biological Sciences of Chinese Academy of Sciences and SJTU-SM, Shanghai 200025, China. -

PLRG1 Antibody Cat
PLRG1 Antibody Cat. No.: 46-203 PLRG1 Antibody 46-203 (5ug/ml) staining of paraffin embedded Human Prostate. Steamed antigen retrieval with citrate buffer pH 6, AP-staining. Specifications HOST SPECIES: Goat SPECIES REACTIVITY: Human, Mouse HOMOLOGY: Expected Species Reactivity based on sequence homology: Rat, Dog, Pig, Cow IMMUNOGEN: The immunogen for this antibody is: C-PVSWKPEIIKRKRF TESTED APPLICATIONS: ELISA, IHC, WB Peptide ELISA: antibody detection limit dilution 1:16000.Western Blot:Approx 60kDa band observed in nuclear lysates of cell lines HEK293 and NIH3T3 (calculated MW of 57.2kDa according to Human NP_002660.1 and 56.9kDa according to Mouse APPLICATIONS: NP_058064.2). Recommended concentration: 0.1-0.3ug/ml. Primary incubation was 1 hour.Immunohistochemistry:Paraffin embedded Human Prostate. Recommended concentration: 5ug/ml. September 28, 2021 1 https://www.prosci-inc.com/plrg1-antibody-46-203.html This antibody is expected to recognise isoform 1 (NP_002660.1) and isoform 2 SPECIFICITY: (NP_001188493.1). POSITIVE CONTROL: 1) Cat. No. 1405 - Mouse Kidney Tissue Lysate Properties Purified from goat serum by ammonium sulphate precipitation followed by antigen PURIFICATION: affinity chromatography using the immunizing peptide. CLONALITY: Polyclonal CONJUGATE: Unconjugated PHYSICAL STATE: Liquid Supplied at 0.5 mg/ml in Tris saline, 0.02% sodium azide, pH7.3 with 0.5% bovine serum BUFFER: albumin. Aliquot and store at -20°C. Minimize freezing and thawing. CONCENTRATION: 500 ug/mL STORAGE CONDITIONS: Aliquot and store at -20˚C. -
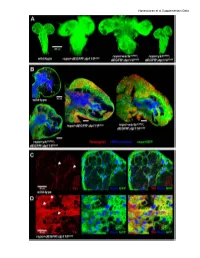
Supplementary Data Vigneswaran Et Al Supplementary Data
Vigneswaran et al Supplementary Data Vigneswaran et al Supplementary Data Figure S1: Yki is required for EGFR-PI3K-driven glial neoplasia in Drosophila (A) Optical projections of whole brain-nerve cord complexes from 3rd instar larvae approximately 130 hrs old. Dorsal view; anterior up. CD8-GFP (green) labels glial cell bodies. Compared to repo>dEGFRλ;dp110CAAX, warts knockdown (repo>wartsdsRNA; dEGFRλ;dp110CAAX) increased neoplastic brain overgrowth and yki knockdown (repo>ykidsRNA;dEGFRλ;dp110CAAX) decreased neoplastic brain overgrowth. (B) 3 µm optical projections of brain hemispheres, age-matched 3rd instar larvae. Frontal sections; anterior up; midline to left. Repo (red) labels glial cell nuclei; CD8-GFP (green) labels glial cell bodies; anti-HRP (blue) counter-stains for neurons and neuropil. (middle) repo>dEGFRλ;dp110CAAX showed increased glial cell numbers (red nuclei) compared to (upper left) wild-type. Compared to repo>dEGFRλ;dp110CAAX, (right) warts knockdown increased neoplastic glial cell numbers (red nuclei), whereas (lower left) yki knockdown reduced neoplastic glial cell numbers (red nuclei). (C, D) Low levels of Yki protein (red) was observed in wild-type central brain glia (white arrows, left panel in C) compared to high levels of cytoplasmic and nuclear Yki protein in dEGFRλ;dp110CAAX neoplastic glia (white arrows, left panel in D); Repo (blue) labels glial cell nuclei; CD8-GFP (green) labels glial cell bodies. Vigneswaran et al Supplementary Data Figure S2: YAP/TAZ expression confined to RTK-amplified tuMor cells and Maintained in patient-derived xenografts (A) On the left, immunohistochemical (IHC) staining in representative normal brain parenchyma in the cortex where YAP expression and TAZ expression was limited to vascular cells and was not detectable in normal neuronal and glial cells. -

Enhancement of RNA Splicing by the Nutrient-Regulated Splicing Factor SRSF3
Graduate Theses, Dissertations, and Problem Reports 2015 Enhancement of RNA Splicing by the Nutrient-Regulated Splicing Factor SRSF3 Amanda L. Suchanek Follow this and additional works at: https://researchrepository.wvu.edu/etd Recommended Citation Suchanek, Amanda L., "Enhancement of RNA Splicing by the Nutrient-Regulated Splicing Factor SRSF3" (2015). Graduate Theses, Dissertations, and Problem Reports. 6740. https://researchrepository.wvu.edu/etd/6740 This Dissertation is protected by copyright and/or related rights. It has been brought to you by the The Research Repository @ WVU with permission from the rights-holder(s). You are free to use this Dissertation in any way that is permitted by the copyright and related rights legislation that applies to your use. For other uses you must obtain permission from the rights-holder(s) directly, unless additional rights are indicated by a Creative Commons license in the record and/ or on the work itself. This Dissertation has been accepted for inclusion in WVU Graduate Theses, Dissertations, and Problem Reports collection by an authorized administrator of The Research Repository @ WVU. For more information, please contact [email protected]. Enhancement of RNA Splicing by the Nutrient-Regulated Splicing Factor SRSF3. Amanda L. Suchanek Dissertation submitted to the School of Medicine at West Virginia University in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biochemistry & Molecular Biology Committee Members Lisa Salati, Ph.D., Chair John Hollander, Ph.D. J. Michael Ruppert, M.D., Ph.D. Maxim Sokolov, Ph.D. Yehenew Agazie, Ph.D. Graduate Program in Biochemistry West Virginia University School of Medicine Morgantown, West Virginia 2015 Keywords: SRSF3, G6PD, RNA splicing, hepatocytes, adenovirus, intron retention © 2015 Amanda Suchanek ABSTRACT Enhancement of RNA Splicing by the Nutrient Regulated Splicing Factor, SRSF3 Amanda L.