Poaceae), and the Interrelation of Whole-Genome Duplication and Evolutionary Radiations in This Grass Tribe
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Environmental Weeds of Coastal Plains and Heathy Forests Bioregions of Victoria Heading in Band
Advisory list of environmental weeds of coastal plains and heathy forests bioregions of Victoria Heading in band b Advisory list of environmental weeds of coastal plains and heathy forests bioregions of Victoria Heading in band Advisory list of environmental weeds of coastal plains and heathy forests bioregions of Victoria Contents Introduction 1 Purpose of the list 1 Limitations 1 Relationship to statutory lists 1 Composition of the list and assessment of taxa 2 Categories of environmental weeds 5 Arrangement of the list 5 Column 1: Botanical Name 5 Column 2: Common Name 5 Column 3: Ranking Score 5 Column 4: Listed in the CALP Act 1994 5 Column 5: Victorian Alert Weed 5 Column 6: National Alert Weed 5 Column 7: Weed of National Significance 5 Statistics 5 Further information & feedback 6 Your involvement 6 Links 6 Weed identification texts 6 Citation 6 Acknowledgments 6 Bibliography 6 Census reference 6 Appendix 1 Environmental weeds of coastal plains and heathy forests bioregions of Victoria listed alphabetically within risk categories. 7 Appendix 2 Environmental weeds of coastal plains and heathy forests bioregions of Victoria listed by botanical name. 19 Appendix 3 Environmental weeds of coastal plains and heathy forests bioregions of Victoria listed by common name. 31 Advisory list of environmental weeds of coastal plains and heathy forests bioregions of Victoria i Published by the Victorian Government Department of Sustainability and Environment Melbourne, March2008 © The State of Victoria Department of Sustainability and Environment 2009 This publication is copyright. No part may be reproduced by any process except in accordance with the provisions of the Copyright Act 1968. -

Jervis Bay Territory Page 1 of 50 21-Jan-11 Species List for NRM Region (Blank), Jervis Bay Territory
Biodiversity Summary for NRM Regions Species List What is the summary for and where does it come from? This list has been produced by the Department of Sustainability, Environment, Water, Population and Communities (SEWPC) for the Natural Resource Management Spatial Information System. The list was produced using the AustralianAustralian Natural Natural Heritage Heritage Assessment Assessment Tool Tool (ANHAT), which analyses data from a range of plant and animal surveys and collections from across Australia to automatically generate a report for each NRM region. Data sources (Appendix 2) include national and state herbaria, museums, state governments, CSIRO, Birds Australia and a range of surveys conducted by or for DEWHA. For each family of plant and animal covered by ANHAT (Appendix 1), this document gives the number of species in the country and how many of them are found in the region. It also identifies species listed as Vulnerable, Critically Endangered, Endangered or Conservation Dependent under the EPBC Act. A biodiversity summary for this region is also available. For more information please see: www.environment.gov.au/heritage/anhat/index.html Limitations • ANHAT currently contains information on the distribution of over 30,000 Australian taxa. This includes all mammals, birds, reptiles, frogs and fish, 137 families of vascular plants (over 15,000 species) and a range of invertebrate groups. Groups notnot yet yet covered covered in inANHAT ANHAT are notnot included included in in the the list. list. • The data used come from authoritative sources, but they are not perfect. All species names have been confirmed as valid species names, but it is not possible to confirm all species locations. -
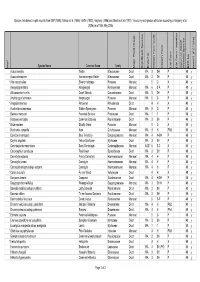
BFS048 Site Species List
Species lists based on plot records from DEP (1996), Gibson et al. (1994), Griffin (1993), Keighery (1996) and Weston et al. (1992). Taxonomy and species attributes according to Keighery et al. (2006) as of 16th May 2005. Species Name Common Name Family Major Plant Group Significant Species Endemic Growth Form Code Growth Form Life Form Life Form - aquatics Common SSCP Wetland Species BFS No kens01 (FCT23a) Wd? Acacia sessilis Wattle Mimosaceae Dicot WA 3 SH P 48 y Acacia stenoptera Narrow-winged Wattle Mimosaceae Dicot WA 3 SH P 48 y * Aira caryophyllea Silvery Hairgrass Poaceae Monocot 5 G A 48 y Alexgeorgea nitens Alexgeorgea Restionaceae Monocot WA 6 S-R P 48 y Allocasuarina humilis Dwarf Sheoak Casuarinaceae Dicot WA 3 SH P 48 y Amphipogon turbinatus Amphipogon Poaceae Monocot WA 5 G P 48 y * Anagallis arvensis Pimpernel Primulaceae Dicot 4 H A 48 y Austrostipa compressa Golden Speargrass Poaceae Monocot WA 5 G P 48 y Banksia menziesii Firewood Banksia Proteaceae Dicot WA 1 T P 48 y Bossiaea eriocarpa Common Bossiaea Papilionaceae Dicot WA 3 SH P 48 y * Briza maxima Blowfly Grass Poaceae Monocot 5 G A 48 y Burchardia congesta Kara Colchicaceae Monocot WA 4 H PAB 48 y Calectasia narragara Blue Tinsel Lily Dasypogonaceae Monocot WA 4 H-SH P 48 y Calytrix angulata Yellow Starflower Myrtaceae Dicot WA 3 SH P 48 y Centrolepis drummondiana Sand Centrolepis Centrolepidaceae Monocot AUST 6 S-C A 48 y Conostephium pendulum Pearlflower Epacridaceae Dicot WA 3 SH P 48 y Conostylis aculeata Prickly Conostylis Haemodoraceae Monocot WA 4 H P 48 y Conostylis juncea Conostylis Haemodoraceae Monocot WA 4 H P 48 y Conostylis setigera subsp. -

Checklist of the Vascular Plants of Redwood National Park
Humboldt State University Digital Commons @ Humboldt State University Botanical Studies Open Educational Resources and Data 9-17-2018 Checklist of the Vascular Plants of Redwood National Park James P. Smith Jr Humboldt State University, [email protected] Follow this and additional works at: https://digitalcommons.humboldt.edu/botany_jps Part of the Botany Commons Recommended Citation Smith, James P. Jr, "Checklist of the Vascular Plants of Redwood National Park" (2018). Botanical Studies. 85. https://digitalcommons.humboldt.edu/botany_jps/85 This Flora of Northwest California-Checklists of Local Sites is brought to you for free and open access by the Open Educational Resources and Data at Digital Commons @ Humboldt State University. It has been accepted for inclusion in Botanical Studies by an authorized administrator of Digital Commons @ Humboldt State University. For more information, please contact [email protected]. A CHECKLIST OF THE VASCULAR PLANTS OF THE REDWOOD NATIONAL & STATE PARKS James P. Smith, Jr. Professor Emeritus of Botany Department of Biological Sciences Humboldt State Univerity Arcata, California 14 September 2018 The Redwood National and State Parks are located in Del Norte and Humboldt counties in coastal northwestern California. The national park was F E R N S established in 1968. In 1994, a cooperative agreement with the California Department of Parks and Recreation added Del Norte Coast, Prairie Creek, Athyriaceae – Lady Fern Family and Jedediah Smith Redwoods state parks to form a single administrative Athyrium filix-femina var. cyclosporum • northwestern lady fern unit. Together they comprise about 133,000 acres (540 km2), including 37 miles of coast line. Almost half of the remaining old growth redwood forests Blechnaceae – Deer Fern Family are protected in these four parks. -

Digitalcommons@University of Nebraska - Lincoln
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln U.S. Department of Agriculture: Agricultural Publications from USDA-ARS / UNL Faculty Research Service, Lincoln, Nebraska 1996 Bromegrasses Kenneth P. Vogel University of Nebraska-Lincoln, [email protected] K. J. Moore Iowa State University Lowell E. Moser University of Nebraska-Lincoln, [email protected] Follow this and additional works at: https://digitalcommons.unl.edu/usdaarsfacpub Vogel, Kenneth P.; Moore, K. J.; and Moser, Lowell E., "Bromegrasses" (1996). Publications from USDA- ARS / UNL Faculty. 2097. https://digitalcommons.unl.edu/usdaarsfacpub/2097 This Article is brought to you for free and open access by the U.S. Department of Agriculture: Agricultural Research Service, Lincoln, Nebraska at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Publications from USDA-ARS / UNL Faculty by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Published 1996 17 Bromegrasses1 K.P. VOGEL USDA-ARS Lincoln, Nebraska K.J.MOORE Iowa State University Ames, Iowa LOWELL E. MOSER University of Nebraska Lincoln, Nebraska The bromegrasses belong to the genus Bromus of which there are some 100 spe cies (Gould & Shaw, 1983). The genus includes both annual and perennial cool season species adapted to temperate climates. Hitchcock (1971) described 42 bro megrass species found in the USA and Canada of which 22 were native (Gould & Shaw, 1983). Bromus is the Greek word for oat and refers to the panicle inflo rescence characteristic of the genus. The bromegrasses are C3 species (Krenzer et aI., 1975; Waller & Lewis, 1979). Of all the bromegrass species, only two are cultivated for permanent pas tures to any extent in North America. -

BSBI News Index 121-130 ABC 8Pt FINAL
BSBI News INDEX to Nos 121 – 130 September 2012 to September 2015 Compiled by GWYNN ELLIS ISSN 2397-8813 1 GUIDE TO THE INDEX ABBREVIATIONS AEM Annual Exhibition Meeting Illus. Illustration AGM Annual General Meeting Infl. Inflorescence ASM Annual Summer Meeting Lvs Leaves cf. confer (compare) photo © photo copyright holder congrats congratulations Rev. Review CS Colour Section Rpt Report del. delineavit (drawn) s.l. sensu lato (broad sense) Descr. Description s.s. sensu stricto (narrow sense) Diag. Diagram v.c. vice-county Exbn Exhibition v.cc. vice-counties Exbt Exhibit (♀) female parent Fld Mtg Rpt Field Meeting Report (♂) male parent Fls Flowers ACKNOWLEDGEMENTS: The compiler wishes to thank David Pearman for much helpful advice and for scrutinising the final text. However, responsibility for checking the index and its final form rests solely with the compiler. BOOKS et al. are italicised as are Periodicals and scientific names COLOUR PAGES: In the index all colour page numbers are distinguished by being underlined with the cover pages enclosed in square brackets [ ]. The front cover and inside front cover are numbered [i] and [ii] respectively while the inside back and back cover pages are numbered according to the number of pages, thus with an issue of 76 pages the inside back cover is [77] and the back cover [78]. Colour Section plates are numbered CS1, CS2, CS3, CS4. Photographers are now indexed by name with the qualification (photo ©) COMPILATION: Using the original text on computer, the entries for each issue were generated by deleting all unwanted text. After checking, the entries were then sorted into alphabetical order, condensed, and finally output as pdf files for the Printer. -

Notes on Grasses (Poaceae) for the Flora of China, VI. New Combinations in Stipeae and Anthoxanthum Author(S): Sylvia M
Notes on Grasses (Poaceae) for the Flora of China, VI. New Combinations in Stipeae and Anthoxanthum Author(s): Sylvia M. Phillips and Wu Zhen-Lan Source: Novon, Vol. 15, No. 3 (Sep., 2005), pp. 474-476 Published by: Missouri Botanical Garden Press Stable URL: http://www.jstor.org/stable/3393497 Accessed: 21-07-2016 02:11 UTC Your use of the JSTOR archive indicates your acceptance of the Terms & Conditions of Use, available at http://about.jstor.org/terms JSTOR is a not-for-profit service that helps scholars, researchers, and students discover, use, and build upon a wide range of content in a trusted digital archive. We use information technology and tools to increase productivity and facilitate new forms of scholarship. For more information about JSTOR, please contact [email protected]. Missouri Botanical Garden Press is collaborating with JSTOR to digitize, preserve and extend access to Novon This content downloaded from 159.226.89.2 on Thu, 21 Jul 2016 02:11:29 UTC All use subject to http://about.jstor.org/terms Notes on Grasses (Poaceae) for the Flora of China, VI. New Combinations in Stipeae and Anthoxanthum S+-lvia;V. Phillips Herbarium, Royal Botanic Gardens, Keu. Surrey TB79 3ABt United Kingdom. [email protected] Wu Zhen-LvIrl Herbarium, Northwest Plateau Institute of Biolog!. \(aclemia Sinica, 78 Xiguan Street, Xining, Qinghai 810()01. China ABSTRACT. When Or^zopsis is e onfined to the tv^)e gelleIic reassessmellt for North America and w-ith species, Chinese species are placed in Achrlathe- ollal-)oIatoIs folloss-ed this uith a report on molec- rum and Piptatherum. -

Poaceae: Pooideae) Based on Plastid and Nuclear DNA Sequences
d i v e r s i t y , p h y l o g e n y , a n d e v o l u t i o n i n t h e monocotyledons e d i t e d b y s e b e r g , p e t e r s e n , b a r f o d & d a v i s a a r h u s u n i v e r s i t y p r e s s , d e n m a r k , 2 0 1 0 Phylogenetics of Stipeae (Poaceae: Pooideae) Based on Plastid and Nuclear DNA Sequences Konstantin Romaschenko,1 Paul M. Peterson,2 Robert J. Soreng,2 Núria Garcia-Jacas,3 and Alfonso Susanna3 1M. G. Kholodny Institute of Botany, Tereshchenkovska 2, 01601 Kiev, Ukraine 2Smithsonian Institution, Department of Botany MRC-166, National Museum of Natural History, P.O. Box 37012, Washington, District of Columbia 20013-7012 USA. 3Laboratory of Molecular Systematics, Botanic Institute of Barcelona (CSIC-ICUB), Pg. del Migdia, s.n., E08038 Barcelona, Spain Author for correspondence ([email protected]) Abstract—The Stipeae tribe is a group of 400−600 grass species of worldwide distribution that are currently placed in 21 genera. The ‘needlegrasses’ are char- acterized by having single-flowered spikelets and stout, terminally-awned lem- mas. We conducted a molecular phylogenetic study of the Stipeae (including all genera except Anemanthele) using a total of 94 species (nine species were used as outgroups) based on five plastid DNA regions (trnK-5’matK, matK, trnHGUG-psbA, trnL5’-trnF, and ndhF) and a single nuclear DNA region (ITS). -

Vascular Plants and a Brief History of the Kiowa and Rita Blanca National Grasslands
United States Department of Agriculture Vascular Plants and a Brief Forest Service Rocky Mountain History of the Kiowa and Rita Research Station General Technical Report Blanca National Grasslands RMRS-GTR-233 December 2009 Donald L. Hazlett, Michael H. Schiebout, and Paulette L. Ford Hazlett, Donald L.; Schiebout, Michael H.; and Ford, Paulette L. 2009. Vascular plants and a brief history of the Kiowa and Rita Blanca National Grasslands. Gen. Tech. Rep. RMRS- GTR-233. Fort Collins, CO: U.S. Department of Agriculture, Forest Service, Rocky Mountain Research Station. 44 p. Abstract Administered by the USDA Forest Service, the Kiowa and Rita Blanca National Grasslands occupy 230,000 acres of public land extending from northeastern New Mexico into the panhandles of Oklahoma and Texas. A mosaic of topographic features including canyons, plateaus, rolling grasslands and outcrops supports a diverse flora. Eight hundred twenty six (826) species of vascular plant species representing 81 plant families are known to occur on or near these public lands. This report includes a history of the area; ethnobotanical information; an introductory overview of the area including its climate, geology, vegetation, habitats, fauna, and ecological history; and a plant survey and information about the rare, poisonous, and exotic species from the area. A vascular plant checklist of 816 vascular plant taxa in the appendix includes scientific and common names, habitat types, and general distribution data for each species. This list is based on extensive plant collections and available herbarium collections. Authors Donald L. Hazlett is an ethnobotanist, Director of New World Plants and People consulting, and a research associate at the Denver Botanic Gardens, Denver, CO. -

Viruses Virus Diseases Poaceae(Gramineae)
Viruses and virus diseases of Poaceae (Gramineae) Viruses The Poaceae are one of the most important plant families in terms of the number of species, worldwide distribution, ecosystems and as ingredients of human and animal food. It is not surprising that they support many parasites including and more than 100 severely pathogenic virus species, of which new ones are being virus diseases regularly described. This book results from the contributions of 150 well-known specialists and presents of for the first time an in-depth look at all the viruses (including the retrotransposons) Poaceae(Gramineae) infesting one plant family. Ta xonomic and agronomic descriptions of the Poaceae are presented, followed by data on molecular and biological characteristics of the viruses and descriptions up to species level. Virus diseases of field grasses (barley, maize, rice, rye, sorghum, sugarcane, triticale and wheats), forage, ornamental, aromatic, wild and lawn Gramineae are largely described and illustrated (32 colour plates). A detailed index Sciences de la vie e) of viruses and taxonomic lists will help readers in their search for information. Foreworded by Marc Van Regenmortel, this book is essential for anyone with an interest in plant pathology especially plant virology, entomology, breeding minea and forecasting. Agronomists will also find this book invaluable. ra The book was coordinated by Hervé Lapierre, previously a researcher at the Institut H. Lapierre, P.-A. Signoret, editors National de la Recherche Agronomique (Versailles-France) and Pierre A. Signoret emeritus eae (G professor and formerly head of the plant pathology department at Ecole Nationale Supérieure ac Agronomique (Montpellier-France). Both have worked from the late 1960’s on virus diseases Po of Poaceae . -

TESIS Caracterización Ambiental De Los Ecosistemas, Zonas De Vida Y
UNIVERSIDAD NACIONAL DANIEL ALCIDES CARRIÓN FACULTAD DE INGENIERÍA ESCUELA DE FORMACIÓN PROFESIONAL DE INGENIERÍA AMBIENTAL TESIS Caracterización Ambiental De Los Ecosistemas, Zonas De Vida Y Vegetación Natural De La Provincia De Pasco Para optar el título profesional de: Ingeniero Ambiental Autor: Bach. Yelitza Orializ VILLEGAS ALANIA Asesor: Ing. Anderson MARCELO MANRIQUE Cerro de Pasco – Perú – 2019 UNIVERSIDAD NACIONAL DANIEL ALCIDES CARRIÓN FACULTAD DE INGENIERÍA ESCUELA DE FORMACIÓN PROFESIONAL DE INGENIERÍA AMBIENTAL TESIS Caracterización Ambiental De Los Ecosistemas, Zonas De Vida Y Vegetación Natural De La Provincia De Pasco Sustentada y aprobada ante los miembros del jurado: Mg. Julio Antonio ASTO LIÑAN Mg. Luis Alberto PACHECO PEÑA PRESIDENTE MIEMBRO Mg. David Johnny CUYUBAMBA ZEVALLOS MIEMBRO 2 DEDICATORIA A mi abuelita, madre y hermanos por el apoyo constante, el empeño y la confianza que me dieron durante el desarrollo y culminación de mis estudios A mi alma mater por brindarme la oportunidad de llegar a ser una profesional 3 RECONOCIMIENTO A Dios por permitirnos tener una buena experiencia dentro de la universidad, por permitir convertimos en profesionales en lo que tanto nos apasiona A mi abuelita, madre y hermanos quienes me brindaron su valiosa y desinteresada orientación y guía en la elaboración del presente trabajo de investigación A cada docente que fue parte de este proceso integral de formación profesional, ya que también fueron un apoyo para poder llegar a esta etapa de nuestras vidas Y a todas las personas que en una u otra forma me apoyaron en la realización de este trabajo 4 RESUMEN La superficie de la Provincia de Pasco abarca dos Eco regiones de las 21 identificadas para nuestro país: La puna de los Andes Centrales y la jalca, las cuales involucran grandes paisajes naturales, con características de formaciones vegetales de matorrales y también de praderas abiertas de gramíneas en los “pajonales” altos andinos. -

Mechanical and Thermal Properties of Insulating Sustainable Mortars with Ampelodesmos Mauritanicus and Pennisetum Setaceum Plants As Aggregates
applied sciences Article Mechanical and Thermal Properties of Insulating Sustainable Mortars with Ampelodesmos mauritanicus and Pennisetum setaceum Plants as Aggregates Dionisio Badagliacco 1 , Carmelo Sanfilippo 1,*, Bartolomeo Megna 1,2 , Tommaso La Mantia 3 and Antonino Valenza 1 1 Department of Engineering, University of Palermo, 90128 Palermo, Italy; [email protected] (D.B.); [email protected] (B.M.); [email protected] (A.V.) 2 INSTM Research Unity of Palermo, 90128 Palermo, Italy 3 Department of Agricultural, Food and Forest Sciences, University of Palermo, 90128 Palermo, Italy; [email protected] * Correspondence: carmelo.sanfi[email protected] Abstract: The use of natural fibers in cement composites is a widening research field as their application can enhance the mechanical and thermal behavior of cement mortars and limit their carbon footprint. In this paper, two different wild grasses, i.e., Ampelodesmos mauritanicus, also called diss, and Pennisetum setaceum, also known as crimson fountaingrass, are used as a source of natural aggregates for cement mortars. The main purpose is to assess the possibility of using the more invasive crimson fountaingrass in place of diss in cement-based vegetable concrete. The two plant fibers have been characterized by means of scanning electron microscopy (SEM), helium Citation: Badagliacco, D.; Sanfilippo, picnometry and thermogravimetric analysis. Moreover, the thermal conductivity of fiber panels has C.; Megna, B.; La Mantia, T.; Valenza, been measured. Mortars samples have been prepared using untreated, boiled and Polyethylene glycol A. Mechanical and Thermal 4000 (PEG) treated fibers. The mechanical characterization has been performed by means of three Properties of Insulating Sustainable point bending and compression tests.