The Complete Mitochondrial Genome of the Chinese Hook Snout
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Biogeography and Evolution of the Carassius Auratus-Complex in East
Takada et al. BMC Evolutionary Biology 2010, 10:7 http://www.biomedcentral.com/1471-2148/10/7 RESEARCH ARTICLE Open Access Biogeography and evolution of the Carassius auratus-complex in East Asia Mikumi Takada1,2*, Katsunori Tachihara1, Takeshi Kon2, Gunji Yamamoto2, Kei’ichiro Iguchi3, Masaki Miya4, Mutsumi Nishida2 Abstract Background: Carassius auratus is a primary freshwater fish with bisexual diploid and unisexual gynogenetic triploid lineages. It is distributed widely in Eurasia and is especially common in East Asia. Although several genetic studies have been conducted on C. auratus, they have not provided clear phylogenetic and evolutionary descriptions of this fish, probably due to selection bias in sampling sites and the DNA regions analysed. As the first step in clarifying the evolutionary entity of the world’s Carassius fishes, we attempted to clarify the phylogeny of C. auratus populations distributed in East Asia. Results: We conducted a detailed analysis of a large dataset of mitochondrial gene sequences [CR, 323 bp, 672 sequences (528 sequenced + 144 downloaded); CR + ND4 + ND5 + cyt b, 4669 bp in total, 53 sequences] obtained from C. auratus in East Asia. Our phylogeographic analysis revealed two superlineages, one distributed mainly among the Japanese main islands and the other in various regions in and around the Eurasian continent, including the Ryukyus and Taiwan. The two superlineages include seven lineages with high regional specificity that are composed of endemic populations indigenous to each region. The divergence time of the seven lineages was estimated to be 0.2 million years ago (Mya) by a fossil-based method and 1.0-1.9 Mya by the molecular clock method. -
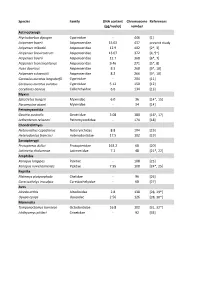
Table S2.Xlsx
Species Family DNA content Chromosome References (pg/nuclei) number Actinopterygii Ptychobarbus dipogon Cyprinidae ‐ 446 [1] Acipenser baerii Acipenseridae 15.02 437 present study Acipenser mikadoi Acipenseridae 12.9 402 [2*, 3] Acipenser brevirostrum Acipenseridae 13.07 372 [4, 5*] Acipenser baerii Acipenseridae 12.7 368 [6*, 7] Acipenser transmontanus Acipenseridae 9.46 271 [5*, 8] Huso dauricus Acipenseridae 8.3 268 [9*, 10] Acipenser schrenckii Acipenseridae 8.2 266 [9*, 10] Carassius auratus langsdorfii Cyprinidae ‐ 204 [11] Carassius auratus auratus Cyprinidae 5.12 150 [12] Corydoras aeneus Callichthyidae 6.6 134 [13] Myxini Eptatretus burgeri Myxinidae 6.0 36 [14*, 15] Paramyxine atami Myxinidae ‐ 34 [14] Petromyzontida Geotria australis Geotriidae 3.08 180 [16*, 17] Lethenteron reissneri Petromyzontidae ‐ 174 [18] Chondrichthyes Notorynchus cepedianus Notorynchidae 8.8 104 [19] Heterodontus francisci Heterodontidae 17.5 102 [19] Sarcopterygii Protopterus dolloi Protopteridae 163.2 68 [20] Latimeria chalumnae Latimeriidae 7.2 48 [21*, 22] Amphibia Xenopus longipes Pipidae ‐ 108 [23] Xenopus ruwenzoriensis Pipidae 7.95 108 [24*, 25] Reptilia Platemys platycephala Chelidae ‐ 96 [26] Carettochelys insculpta Carettochelyidae ‐ 68 [27] Aves Alcedo atthis Alcedinidae 2.8 138 [28, 29*] Upupa epops Upupidae 2.56 126 [28, 30*] Mammalia Tympanoctomys barrerae Octodontidae 16.8 102 [31, 32*] Ichthyomys pittieri Cricetidae ‐ 92 [33] References 1. Yu XY, Yu XJ. A schizothoracin fish species, Diptychus dipogon, with very high number of chromosomes Chrom Inform Serv. 1990;48:17-8. 2. Zhou H, Fujimoto T, Adachi S, Yamaha E, Arai K. Genome size variation estimated by flow cytometry in Acipenser mikadoi, Huso dauricus in relation to other species of Acipenseriformes. -

Strategies for Conservation and Restoration of Freshwater Fish Species in Korea
KOREAN JOURNAL OF ICHTHYOLOGY, Vol. 21 Supplement, 29-37, July 2009 Received : April 22, 2009 ISSN: 1225-8598 Revised : June 6, 2009 Accepted : June 20, 2009 Strategies for Conservation and Restoration of Freshwater Fish Species in Korea By Eon-Jong Kang*, In-Chul Bang1 and Hyun Yang2 Inland Aquaculture Research Center, National Fisheries Research and Development Institute, Busan 619-902, Korea 1Department of Marine Biotechnology, Soonchunhyang University, Asan 336-745, Korea 2Institute of Biodiversity Research, Jeonju 561-211, Korea ABSTRACT The tiny fragment of freshwater body is providing home for huge biodiversity and resour- ces for the existence of human. The competing demand for freshwater have been increased rapidly and it caused the declination of biodiversity in recent decades. Unlike the natural process of extinction in gradual progress, the current species extinction is accelerated by human activity. As a result many fish species are already extinct or alive only in captivity in the world and about fifty eight animal species are in endangered in Korea including eighteen freshwater species. Conservation of biodiversity is the pro- cess by which the prevention of loss or damage is attained, and is often associated with management of the natural environment. The practical action is classified into in-situ, or ex-situ depending on the location of the conservation effort. Recovery means the process by which the status of endangerment is improved to persist in the wild by re-introduction of species from ex-situ conservation population into nature or translocation of some population. However there are a lot of restrictions to complete it and successful results are known very rare in case. -

13. Kneriidae = 13. Kneriidae
13. KNERIIDAE Robert C. SCHELLY Four anatomically disparate genera (Kneria, Parakneria, Grasseichthys and Cromeria) and 24 species are currently recognised within the Kneriidae. Kneriids are anotophysian members of the Ostariophysi and lack a Weberian apparatus. They are found in tropical Africa (including the Nile basin), mostly in flowing waters. Two of the four genera are represented in Lower Guinea: Grasseichthys and Parakneria. Parakneria are readily distinguished from Grasseichthys by their cylindrical body with a flat ventral surface, inferior mouth with keratinized ridges in place of teeth, numerous small scales and large, paddle- shaped pectoral fins positioned abdominally. KEY Very small (20 mm or less in SL) and larval in appearance, laterally TO GENERA compressed, scaleless, with terminal mouth ..... Grasseichthys Adults reach 60 mm SL or more, torpedo-shaped with flat ventral surface, body covered with small cycloid scales, mouth inferior .............................................................................. Parakneria Genus Grasseichthys Géry, 1964 Grasseichthys are miniaturized, scaleless fish with a small, terminal and toothless mouth and small gill openings. Because of its ‘larval appearance’, small size and lack of skeletal ossification, Grasseichthys has been called a paedomorphic kneriid. Grasseichthys gabonensis and ventral edge (ventral extension Géry, 1964 is more developed). Caudal fin forked, with 16 rays, including an unbranched Description: small, elongate, relatively ray on the dorsal and ventral edge. shallow-bodied (BD 17.9-20.0% SL Fin spines lacking. Dorsal III, 4-5; in females and 15.2-17.9% SL in males) anal III, 6-7. Vertebrae, 35-36. and laterally compressed. Head small Two branchiostegal rays. Body scaleless (HL 18.0-22.7% SL), with small, and semi-translucent, with myomeres terminal mouth and large eyes. -

PDF Download
Original Article PNIE 2021;2(1):53-61 https://doi.org/10.22920/PNIE.2021.2.1.53 pISSN 2765-2203, eISSN 2765-2211 Microhabitat Characteristics Determine Fish Community Structure in a Small Stream (Yudeung Stream, South Korea) Jong-Yun Choi , Seong-Ki Kim , Jeong-Cheol Kim , Hyeon-Jeong Lee , Hyo-Jeong Kwon , Jong-Hak Yun* National Institute of Ecology, Seocheon, Korea ABSTRACT Distribution of fish community depends largely on environmental disturbance such as habitat change. In this study, we evaluated the impact of environmental variables and microhabitat patch types on fish distribution in Yudeung Stream at 15 sites between early May and late June 2019. We used non-metric multidimensional scaling to examine the distribution patterns of fish in each site. Gnathopogon strigatus, Squalidus gracilis majimae, Zacco koreanus, and Zacco platypus were associated with riffle and boulder areas, whereas Iksookimia koreensis, Acheilognathus koreensis, Coreoleuciscus splendidus, Sarcocheilichthys nigripinnis morii, and Odontobutis interrupta were associated with large shallow areas. In contrast, Cyprinus carpio, Carassius auratus, Lepomis macrochirus, and Micropterus salmoides were found at downstream sites associated with large pool areas, sandy/clay-bottomed areas, and vegetated areas. On the basis of these results, we suggest that microhabitat patch types are important in determining the diversity and abundance of fish communities, since a mosaic of different microhabitats supports diverse fish species. As such, microhabitat patches are key components of freshwater stream ecosystem heterogeneity, and a suitable patch composition in stream construction or restoration schemes will support ecologically healthy food webs. Keywords: Aquatic macrophytes, Microhabitat, Pool, Riffle, River continuum, Zacco koreanus Introduction 2012). -

Constraints on the Timescale of Animal Evolutionary History
Palaeontologia Electronica palaeo-electronica.org Constraints on the timescale of animal evolutionary history Michael J. Benton, Philip C.J. Donoghue, Robert J. Asher, Matt Friedman, Thomas J. Near, and Jakob Vinther ABSTRACT Dating the tree of life is a core endeavor in evolutionary biology. Rates of evolution are fundamental to nearly every evolutionary model and process. Rates need dates. There is much debate on the most appropriate and reasonable ways in which to date the tree of life, and recent work has highlighted some confusions and complexities that can be avoided. Whether phylogenetic trees are dated after they have been estab- lished, or as part of the process of tree finding, practitioners need to know which cali- brations to use. We emphasize the importance of identifying crown (not stem) fossils, levels of confidence in their attribution to the crown, current chronostratigraphic preci- sion, the primacy of the host geological formation and asymmetric confidence intervals. Here we present calibrations for 88 key nodes across the phylogeny of animals, rang- ing from the root of Metazoa to the last common ancestor of Homo sapiens. Close attention to detail is constantly required: for example, the classic bird-mammal date (base of crown Amniota) has often been given as 310-315 Ma; the 2014 international time scale indicates a minimum age of 318 Ma. Michael J. Benton. School of Earth Sciences, University of Bristol, Bristol, BS8 1RJ, U.K. [email protected] Philip C.J. Donoghue. School of Earth Sciences, University of Bristol, Bristol, BS8 1RJ, U.K. [email protected] Robert J. -

Morphologies and Population Genetic Structures of the Eight-Barbel Loach
Ichthyological Research https://doi.org/10.1007/s10228-020-00783-1 FULL PAPER Morphologies and population genetic structures of the eight‑barbel loach of the genus Lefua on southern Sakhalin Yoshiyasu Machida1 · Minoru Kanaiwa2 · Sergey V. Shedko3 · Hajime Matsubara4 · Hirozumi Kobayashi5 · Ixchel F. Mandagi5,6 · Akira Ooyagi7 · Kazunori Yamahira5 Received: 7 December 2019 / Revised: 5 August 2020 / Accepted: 6 September 2020 © The Ichthyological Society of Japan 2020 Abstract Coldwater, primary freshwater, fsh such as the eight-barbel loach, Lefua nikkonis, are thought to have colonised Hokkaido from the continental Far East via Sakhalin Island during the Late Pleistocene. Lefua populations have been reported on southern Sakhalin, but detailed morphological and population structure analyses have not yet been completed. This infor- mation is important for reconstructing the colonisation history of L. nikkonis to Hokkaido. In this study, morphological analysis revealed that L. nikkonis and two continental congeners, Lefua pleskei and Lefua costata, are distinguishable from each other, and Lefua collected from southern Sakhalin is morphologically more similar to L. nikkonis. Random forest analysis, a machine learning classifcation method, classifed all Sakhalin individuals as L. nikkonis. Haplotype analysis of mitochondrial DNA sequences revealed that all but one Sakhalin haplotype are shared with L. nikkonis and none is shared with the two continental congeners, which supports the hypothesis that Sakhalin Lefua are L. nikkonis. However, none of the Sakhalin haplotypes was distributed on northern Hokkaido. This discontinuous distribution of haplotypes across Sakhalin and Hokkaido suggests that the Sakhalin Lefua populations are not native. Some of the Sakhalin haplotypes were found only on Hokkaido’s Ishikari River system or the Tokachi River system, suggesting that they originated from these regions. -

Family-Cyprinidae-Gobioninae-PDF
SUBFAMILY Gobioninae Bleeker, 1863 - gudgeons [=Gobiones, Gobiobotinae, Armatogobionina, Sarcochilichthyna, Pseudogobioninae] GENUS Abbottina Jordan & Fowler, 1903 - gudgeons, abbottinas [=Pseudogobiops] Species Abbottina binhi Nguyen, in Nguyen & Ngo, 2001 - Cao Bang abbottina Species Abbottina liaoningensis Qin, in Lui & Qin et al., 1987 - Yingkou abbottina Species Abbottina obtusirostris (Wu & Wang, 1931) - Chengtu abbottina Species Abbottina rivularis (Basilewsky, 1855) - North Chinese abbottina [=lalinensis, psegma, sinensis] GENUS Acanthogobio Herzenstein, 1892 - gudgeons Species Acanthogobio guentheri Herzenstein, 1892 - Sinin gudgeon GENUS Belligobio Jordan & Hubbs, 1925 - gudgeons [=Hemibarboides] Species Belligobio nummifer (Boulenger, 1901) - Ningpo gudgeon [=tientaiensis] Species Belligobio pengxianensis Luo et al., 1977 - Sichuan gudgeon GENUS Biwia Jordan & Fowler, 1903 - gudgeons, biwas Species Biwia springeri (Banarescu & Nalbant, 1973) - Springer's gudgeon Species Biwia tama Oshima, 1957 - tama gudgeon Species Biwia yodoensis Kawase & Hosoya, 2010 - Yodo gudgeon Species Biwia zezera (Ishikawa, 1895) - Biwa gudgeon GENUS Coreius Jordan & Starks, 1905 - gudgeons [=Coripareius] Species Coreius cetopsis (Kner, 1867) - cetopsis gudgeon Species Coreius guichenoti (Sauvage & Dabry de Thiersant, 1874) - largemouth bronze gudgeon [=platygnathus, zeni] Species Coreius heterodon (Bleeker, 1865) - bronze gudgeon [=rathbuni, styani] Species Coreius septentrionalis (Nichols, 1925) - Chinese bronze gudgeon [=longibarbus] GENUS Coreoleuciscus -

Polyphyletic Origin of Ornamental Goldfish
Food and Nutrition Sciences, 2015, 6, 1005-1013 Published Online August 2015 in SciRes. http://www.scirp.org/journal/fns http://dx.doi.org/10.4236/fns.2015.611104 Polyphyletic Origin of Ornamental Goldfish Aleksandr V. Podlesnykh1, Vladimir A. Brykov1,2, Lubov A. Skurikhina1 1Far Eastern Branch of the Russian Academy of Sciences, A. V. Zhirmunsky Institute of Marine Biology, Vladivostok, Russia 2School of Natural Sciences, Far Eastern Federal University, Vladivostok, Russia Email: [email protected] Received 6 May 2015; accepted 17 August 2015; published 20 August 2015 Copyright © 2015 by authors and Scientific Research Publishing Inc. This work is licensed under the Creative Commons Attribution International License (CC BY). http://creativecommons.org/licenses/by/4.0/ Abstract Mitochondrial DNA fragment of cytb was compared in all species Carassius auratus complex and three forms of ornamental goldfish. It is shown that the phylogenetic relationships between com- plex species correspond to the existing views, based on mtDNA data and geographical distribution. All forms of ornamental goldfish have a monophyletic origin from Chinese goldfish C. auratus au- ratus. The analysis showed that three nuclear genes (rps7, GH1 and Rh) in the two forms of orna- mental goldfish (Oriental twintail goldfish and Chinese Ranchu) were almost identical C. auratus auratus genes, while all three gene in another more simple form of goldfish (common goldfish) were highly homologous to carp Cyprinus carpio nuclear genes. The obtained data suggested that in the history of aquarium goldfish breeding occurred the stage of distant hybridization between goldfish and common carp. Subsequently, the nuclear genomes of some ornamental forms could be enriched by goldfish genes (a relatively recent form as Oriental twintail goldfish and Chinese Ranchu) or common carp genes (the simplest and most ancient forms like common goldfish) as a result of multidirectional breeding and selection of aquarium goldfish various forms. -

Jlb Smith Institute of Ichthyology
ISSN 0075-2088 J.L.B. SMITH INSTITUTE OF ICHTHYOLOGY GRAHAMSTOWN, SOUTH AFRICA SPECIAL PUBLICATION No. 56 SCIENTIFIC AND COMMON NAMES OF SOUTHERN AFRICAN FRESHWATER FISHES by Paul H. Skelton November 1993 SERIAL PUBLICATIONS o f THE J.L.B. SMITH INSTITUTE OF ICHTHYOLOGY The Institute publishes original research on the systematics, zoogeography, ecology, biology and conservation of fishes. Manuscripts on ancillary subjects (aquaculture, fishery biology, historical ichthyology and archaeology pertaining to fishes) will be considered subject to the availability of publication funds. Two series are produced at irregular intervals: the Special Publication series and the Ichthyological Bulletin series. Acceptance of manuscripts for publication is subject to the approval of reviewers from outside the Institute. Priority is given to papers by staff of the Institute, but manuscripts from outside the Institute will be considered if they are pertinent to the work of the Institute. Colour illustrations can be printed at the expense of the author. Publications of the Institute are available by subscription or in exchange for publi cations of other institutions. Lists of the Institute’s publications are available from the Publications Secretary at the address below. INSTRUCTIONS TO AUTHORS Manuscripts shorter than 30 pages will generally be published in the Special Publications series; longer papers will be considered for the Ichthyological Bulletin series. Please follow the layout and format of a recent Bulletin or Special Publication. Manuscripts must be submitted in duplicate to the Editor, J.L.B. Smith Institute of Ichthyology, Private Bag 1015, Grahamstown 6140, South Africa. The typescript must be double-spaced throughout with 25 mm margins all round. -

And Intra-Species Replacements in Freshwater Fishes in Japan
G C A T T A C G G C A T genes Article Waves Out of the Korean Peninsula and Inter- and Intra-Species Replacements in Freshwater Fishes in Japan Shoji Taniguchi 1 , Johanna Bertl 2, Andreas Futschik 3 , Hirohisa Kishino 1 and Toshio Okazaki 1,* 1 Graduate School of Agricultural and Life Sciences, The University of Tokyo, 1-1-1, Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan; [email protected] (S.T.); [email protected] (H.K.) 2 Department of Mathematics, Aarhus University, Ny Munkegade, 118, bldg. 1530, 8000 Aarhus C, Denmark; [email protected] 3 Department of Applied Statistics, Johannes Kepler University Linz, Altenberger Str. 69, 4040 Linz, Austria; [email protected] * Correspondence: [email protected] Abstract: The Japanese archipelago is located at the periphery of the continent of Asia. Rivers in the Japanese archipelago, separated from the continent of Asia by about 17 Ma, have experienced an intermittent exchange of freshwater fish taxa through a narrow land bridge generated by lowered sea level. As the Korean Peninsula and Japanese archipelago were not covered by an ice sheet during glacial periods, phylogeographical analyses in this region can trace the history of biota that were, for a long time, beyond the last glacial maximum. In this study, we analyzed the phylogeography of four freshwater fish taxa, Hemibarbus longirostris, dark chub Nipponocypris temminckii, Tanakia ssp. and Carassius ssp., whose distributions include both the Korean Peninsula and Western Japan. We found for each taxon that a small component of diverse Korean clades of freshwater fishes Citation: Taniguchi, S.; Bertl, J.; migrated in waves into the Japanese archipelago to form the current phylogeographic structure of Futschik, A.; Kishino, H.; Okazaki, T. -

PHYLOGENY and ZOOGEOGRAPHY of the SUPERFAMILY COBITOIDEA (CYPRINOIDEI, Title CYPRINIFORMES)
PHYLOGENY AND ZOOGEOGRAPHY OF THE SUPERFAMILY COBITOIDEA (CYPRINOIDEI, Title CYPRINIFORMES) Author(s) SAWADA, Yukio Citation MEMOIRS OF THE FACULTY OF FISHERIES HOKKAIDO UNIVERSITY, 28(2), 65-223 Issue Date 1982-03 Doc URL http://hdl.handle.net/2115/21871 Type bulletin (article) File Information 28(2)_P65-223.pdf Instructions for use Hokkaido University Collection of Scholarly and Academic Papers : HUSCAP PHYLOGENY AND ZOOGEOGRAPHY OF THE SUPERFAMILY COBITOIDEA (CYPRINOIDEI, CYPRINIFORMES) By Yukio SAWADA Laboratory of Marine Zoology, Faculty of Fisheries, Bokkaido University Contents page I. Introduction .......................................................... 65 II. Materials and Methods ............... • • . • . • . • • . • . 67 m. Acknowledgements...................................................... 70 IV. Methodology ....................................•....•.........•••.... 71 1. Systematic methodology . • • . • • . • • • . 71 1) The determinlttion of polarity in the morphocline . • . 72 2) The elimination of convergence and parallelism from phylogeny ........ 76 2. Zoogeographical methodology . 76 V. Comparative Osteology and Discussion 1. Cranium.............................................................. 78 2. Mandibular arch ...................................................... 101 3. Hyoid arch .......................................................... 108 4. Branchial apparatus ...................................•..••......••.. 113 5. Suspensorium.......................................................... 120 6. Pectoral