Diverse RNA Viruses of Arthropod Origin in the Blood of Fruit Bats Suggest a Link Between Bat and Arthropod Viromes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Differential Segregation of Nodaviral Coat Protein and RNA Into Progeny Virions During Mixed Infection with FHV and Nov
Virology 454-455 (2014) 280–290 Contents lists available at ScienceDirect Virology journal homepage: www.elsevier.com/locate/yviro Differential segregation of nodaviral coat protein and RNA into progeny virions during mixed infection with FHV and NoV Radhika Gopal, P. Arno Venter 1, Anette Schneemann n Department of Cell and Molecular Biology, The Scripps Research Institute, La Jolla, CA, USA article info abstract Article history: Nodaviruses are icosahedral viruses with a bipartite, positive-sense RNA genome. The two RNAs are Received 30 December 2013 packaged into a single virion by a poorly understood mechanism. We chose two distantly related Returned to author for revisions nodaviruses, Flock House virus and Nodamura virus, to explore formation of viral reassortants as a 27 January 2014 means to further understand genome recognition and encapsidation. In mixed infections, the viruses Accepted 3 March 2014 were incompatible at the level of RNA replication and their coat proteins segregated into separate Available online 21 March 2014 populations of progeny particles. RNA packaging, on the other hand, was indiscriminate as all four viral Keywords: RNAs were detectable in each progeny population. Consistent with the trans-encapsidation phenotype, Flock House virus fluorescence in situ hybridization of viral RNA revealed that the genomes of the two viruses co-localized Nodamura virus throughout the cytoplasm. Our results imply that nodaviral RNAs lack rigorously defined packaging Mixed infection signals and that co-encapsidation of the viral RNAs does not require a pair of cognate RNA1 and RNA2. Viral assembly & RNA encapsidation 2014 Elsevier Inc. All rights reserved. Viral reassortant Introduction invaginations of the outer membrane of the organelle (Kopek et al., 2007). -
Bulged Stem-Loop
Proc. Nati. Acad. Sci. USA Vol. 89, pp. 11146-11150, December 1992 Biochemisty Evidence that the packaging signal for nodaviral RNA2 is a bulged stem-loop (defecfive-interfering RNA/blpartite genone/RNA p ag) WEIDONG ZHONG, RANJIT DASGUPTA, AND ROLAND RUECKERT* Institute for Molecular Virology and Department of Biochemistry, University of Wisconsin, 1525 Linden Drive, Madison, WI 53706 Communicated by Paul Kaesberg, August 26, 1992 ABSTRACT Flock house virus is an insect virus ging tion initiation site of T3 promoter and the cloned DI DNA to the family Nodaviridae; members of this family are char- through oligonucleotide-directed mutagenesis. Such RNA acterized by a small bipartite positive-stranded RNA genome. transcripts, however, still had four extra nonviral bases atthe The blrger genomic m , RNA1, encodes viral repliation 3' end (4). proteins, whereas the smaller one, RNA2, e coat protein. In Viro Transcription of Cloned FIV DNA. Selected DNA Both RNAsarepa ed in a single particle. A defective- clones were cleaved with the restriction enzyme Xba I, and interferin RNA (DI-634), isolated from a line of DrosophUa the resulting linear DNA templates (0.03 mg/ml) were tran- cells persistently infected with Flock house virus, was used to scribed with T3 RNA polymerase as described by Konarska show that a 32-base regionofRNA2 (bases 186-217) is required et al. (20). One-half millimolar guanosine(5')triphospho- for pcaing into virions. RNA folding analysis predicted that (5')guanosine [G(5')ppp(5')GI was included in the reaction this region forms a stem-loop structure with a 5-base loop and mixture to provide capped transcripts. -

Viral Haemorrhagic Septicaemia Virus (VHSV): on the Search for Determinants Important for Virulence in Rainbow Trout Oncorhynchus Mykiss
Downloaded from orbit.dtu.dk on: Nov 08, 2017 Viral haemorrhagic septicaemia virus (VHSV): on the search for determinants important for virulence in rainbow trout oncorhynchus mykiss Olesen, Niels Jørgen; Skall, H. F.; Kurita, J.; Mori, K.; Ito, T. Published in: 17th International Conference on Diseases of Fish And Shellfish Publication date: 2015 Document Version Publisher's PDF, also known as Version of record Link back to DTU Orbit Citation (APA): Olesen, N. J., Skall, H. F., Kurita, J., Mori, K., & Ito, T. (2015). Viral haemorrhagic septicaemia virus (VHSV): on the search for determinants important for virulence in rainbow trout oncorhynchus mykiss. In 17th International Conference on Diseases of Fish And Shellfish: Abstract book (pp. 147-147). [O-139] Las Palmas: European Association of Fish Pathologists. General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the public portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the public portal If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. DISCLAIMER: The organizer takes no responsibility for any of the content stated in the abstracts. -

Oryzias Latipes)
Betanodavirus infection in the freshwater model fish medaka (Oryzias latipes) Ryo Furusawa, Yasushi Okinaka,* and Toshihiro Nakai Graduate School of Biosphere Science, Hiroshima University, Higashi-hiroshima 739- 8528, Japan æfAuthor for correspondence: Yasushi Okinaka. Telephone: +8 1-82-424-7978. Fax: +81- 82-424-79 1 6. E-mail: [email protected] Running title: Betanodavirus infection in medaka Key words: medaka, betanodavirus, model fish, model virus, freshwater fish Total number of words; text (3532 words), summary (230 words) Total number of figures; 6 figures Total number of tables; 0 table SUMMARY Betanodaviruses, the causal agents of viral nervous necrosis in marine fish, have bipartite positive-sense RNA genomes. Because the genomes are the smallest and simplest among viruses, betanodaviruses are well studied using a genetic engineering system as model viruses, like the cases with the insect viruses, alphanodaviruses, the other members of the family Nodaviridae. However, studies of virus-host interactions have been limited because betanodaviruses basically infect marine fish at early developmental stages (larval and juvenile). These fish are only available for a few months of the year and are not suitable for the construction of a reversed genetics system. To overcome these problems, several freshwater fish species were tested for their susceptibility to betanodaviruses. We have demonstrated that adult medaka (Oryzias latipes), a well-known model fish, is susceptible to both Stripedjack nervous necrosis virus (the type species of the betanodaviruses) and Redspotted grouper nervous necrosis virus which have different host specificity in marine fish species. Infected medaka exhibited erratic swimming and the viruses were specifically localized to the brain, spinal cord, and retina of the infected fish, similar to the pattern of infection in naturally infected marine fish. -

Virus Replication
Introduction • Encompasses > 150 viruses Rhabdoviridae •Rabies –only important human Brian Wells pathogen • One of the most lethal of all infectious diseases History History • Adapted from Latin meaning “to rage” • 1885 – Louis Pasteur – rabies vaccine • Greeks – lyssa – “frenzy” – Attenuated form of virus produced by inoculation of rabbit spinal cord • Rabies represents one of the oldest and most feared diseases • Occurs throughout the world except in Australia, Japan, Great Britain, • Recognized in Egypt before 2300 B.C. and islands such as Hawaii • Well described by Aristotle • “Reportable” disease • Iliad – “canine madness” Taxonomy Viral Structure • 3 Genera – Ephemerovirus, Lyssavirus, • 170 nm x 70 nm Vesiculovirus • Bullet-shaped enveloped virion • Infect vertebrates, invertebrates, and – Glycoprotein peplomers & matrix protein plants under envelope • Genus Lyssavirus comprises rabies • Helical symmetry virus and 3 rabies-like viruses • Linear minus sense ssRNA – 11-12 kb • Each capable of causing rabies-like • Glycoprotein spikes in outer membrane disease in humans bilayer 1 Virus Replication • Receptor-mediated endocytosis • Uncoat and release nucleocapsid into cytoplasm • Production of 5 monocistronic mRNA species - N, P (NS), M, G, L – by L+P viral transcriptase • Each mRNA capped and poly-A’ed • dsRNA replicative intermediate Virus Replication Virus Replication • N+P+L and (-) ssRNA form core • M forms matrix around core • Virus buds from glycoprotein area of plasma membrane and thus acquires its envelope Transmission • Unstable -

Daytime Behaviour of the Grey-Headed Flying Fox Pteropus Poliocephalus Temminck (Pteropodidae: Megachiroptera) at an Autumn/Winter Roost
DAYTIME BEHAVIOUR OF THE GREY-HEADED FLYING FOX PTEROPUS POLIOCEPHALUS TEMMINCK (PTEROPODIDAE: MEGACHIROPTERA) AT AN AUTUMN/WINTER ROOST K.A. CONNELL, U. MUNRO AND F.R. TORPY Connell KA, Munro U and Torpy FR, 2006. Daytime behaviour of the grey-headed flying fox Pteropus poliocephalus Temminck (Pteropodidae: Megachiroptera) at an autumn/winter roost. Australian Mammalogy 28: 7-14. The grey-headed flying fox (Pteropus poliocephalus Temminck) is a threatened large fruit bat endemic to Australia. It roosts in large colonies in rainforest patches, mangroves, open forest, riparian woodland and, as native habitat is reduced, increasingly in vegetation within urban environments. The general biology, ecology and behaviour of this bat remain largely unknown, which makes it difficult to effectively monitor, protect and manage this species. The current study provides baseline information on the daytime behaviour of P. poliocephalus in an autumn/winter roost in urban Sydney, Australia, between April and August 2003. The most common daytime behaviours expressed by the flying foxes were sleeping (most common), grooming, mating/courtship, and wing spreading (least common). Behaviours differed significantly between times of day and seasons (autumn and winter). Active behaviours (i.e., grooming, mating/courtship, wing spreading) occurred mainly in the morning, while sleeping predominated in the afternoon. Mating/courtship and wing spreading were significantly higher in April (reproductive period) than in winter (non-reproductive period). Grooming was the only behaviour that showed no significant variation between sample periods. These results provide important baseline data for future comparative studies on the behaviours of flying foxes from urban and ‘natural’ camps, and the development of management strategies for this species. -

Virus Particle Structures
Virus Particle Structures Virus Particle Structures Palmenberg, A.C. and Sgro, J.-Y. COLOR PLATE LEGENDS These color plates depict the relative sizes and comparative virion structures of multiple types of viruses. The renderings are based on data from published atomic coordinates as determined by X-ray crystallography. The international online repository for 3D coordinates is the Protein Databank (www.rcsb.org/pdb/), maintained by the Research Collaboratory for Structural Bioinformatics (RCSB). The VIPER web site (mmtsb.scripps.edu/viper), maintains a parallel collection of PDB coordinates for icosahedral viruses and additionally offers a version of each data file permuted into the same relative 3D orientation (Reddy, V., Natarajan, P., Okerberg, B., Li, K., Damodaran, K., Morton, R., Brooks, C. and Johnson, J. (2001). J. Virol., 75, 11943-11947). VIPER also contains an excellent repository of instructional materials pertaining to icosahedral symmetry and viral structures. All images presented here, except for the filamentous viruses, used the standard VIPER orientation along the icosahedral 2-fold axis. With the exception of Plate 3 as described below, these images were generated from their atomic coordinates using a novel radial depth-cue colorization technique and the program Rasmol (Sayle, R.A., Milner-White, E.J. (1995). RASMOL: biomolecular graphics for all. Trends Biochem Sci., 20, 374-376). First, the Temperature Factor column for every atom in a PDB coordinate file was edited to record a measure of the radial distance from the virion center. The files were rendered using the Rasmol spacefill menu, with specular and shadow options according to the Van de Waals radius of each atom. -

Journal of Virology
JOURNAL OF VIROLOGY Volume 80 March 2006 No. 6 SPOTLIGHT Articles of Significant Interest Selected from This Issue by 2587–2588 the Editors STRUCTURE AND ASSEMBLY Crystal Structure of the Oligomerization Domain of the Haitao Ding, Todd J. Green, 2808–2814 Phosphoprotein of Vesicular Stomatitis Virus Shanyun Lu, and Ming Luo Subcellular Localization of Hepatitis C Virus Structural Yves Rouille´, Franc¸ois Helle, David 2832–2841 Proteins in a Cell Culture System That Efficiently Replicates Delgrange, Philippe Roingeard, the Virus Ce´cile Voisset, Emmanuelle Blanchard, Sandrine Belouzard, Jane McKeating, Arvind H. Patel, Geert Maertens, Takaji Wakita, Czeslaw Wychowski, and Jean Dubuisson A Small Loop in the Capsid Protein of Moloney Murine Marcy R. Auerbach, Kristy R. 2884–2893 Leukemia Virus Controls Assembly of Spherical Cores Brown, Artem Kaplan, Denise de Las Nueces, and Ila R. Singh Identification of the Nucleocapsid, Tegument, and Envelope Jyh-Ming Tsai, Han-Ching Wang, 3021–3029 Proteins of the Shrimp White Spot Syndrome Virus Virion Jiann-Horng Leu, Andrew H.-J. Wang, Ying Zhuang, Peter J. Walker, Guang-Hsiung Kou, and Chu-Fang Lo GENOME REPLICATION AND REGULATION OF VIRAL GENE EXPRESSION Epitope Mapping of Herpes Simplex Virus Type 2 gH/gL Tina M. Cairns, Marie S. Shaner, Yi 2596–2608 Defines Distinct Antigenic Sites, Including Some Associated Zuo, Manuel Ponce-de-Leon, with Biological Function Isabelle Baribaud, Roselyn J. Eisenberg, Gary H. Cohen, and J. Charles Whitbeck The ␣-TIF (VP16) Homologue (ETIF) of Equine Jens von Einem, Daniel 2609–2620 Herpesvirus 1 Is Essential for Secondary Envelopment Schumacher, Dennis J. O’Callaghan, and Virus Egress and Nikolaus Osterrieder Suppression of Viral RNA Recombination by a Host Chi-Ping Cheng, Elena Serviene, 2631–2640 Exoribonuclease and Peter D. -

The Retromer Is Co-Opted to Deliver Lipid Enzymes for the Biogenesis of Lipid-Enriched Tombusviral Replication Organelles
The retromer is co-opted to deliver lipid enzymes for the biogenesis of lipid-enriched tombusviral replication organelles Zhike Fenga, Jun-ichi Inabaa, and Peter D. Nagya,1 aDepartment of Plant Pathology, University of Kentucky, Lexington, KY 40546 Edited by George E. Bruening, University of California, Davis, CA, and approved November 5, 2020 (received for review July 29, 2020) Biogenesis of viral replication organelles (VROs) is critical for repli- TBSV infections include extensive membrane contact sites (vMCSs) cation of positive-strand RNA viruses. In this work, we demonstrate and harbor numerous spherules (containing VRCs), which are that tomato bushy stunt virus (TBSV) and the closely related carna- vesicle-like invaginations in the peroxisomal membranes (8, 11–13). tion Italian ringspot virus (CIRV) hijack the retromer to facilitate A major gap in our understanding of the biogenesis of VROs, in- building VROs in the surrogate host yeast and in plants. Depletion cluding vMCSs and VRCs, is how the cellular lipid-modifying en- of retromer proteins, which are needed for biogenesis of endosomal zymes are recruited to the sites of viral replication. tubular transport carriers, strongly inhibits the peroxisome-associ- Tombusviruses belong to the large Flavivirus-like supergroup ated TBSV and the mitochondria-associated CIRV replication in yeast that includes important human, animal, and plant pathogens. in planta. and In vitro reconstitution revealed the need for the ret- Tombusviruses have a small single-component (+)RNA genome romer for the full activity of the viral replicase. The viral p33 repli- of ∼4.8 kb that codes for five proteins. Among those, there are cation protein interacts with the retromer complex, including Vps26, two essential replication proteins, namely p33 and p92pol, the Vps29, and Vps35. -

Middle European Euophrys C. L. Koch, 1834 (Araneae: Salticidae)—One, Two Or Three Genera?
1998. P. A. Selden (ed.). Proceedings of the 17th European Colloquium of Arachnology, Edinburgh 1997. Middle European Euophrys C. L. Koch, 1834 (Araneae: Salticidae)—one, two or three genera? Marek Z˙abka1 and Jerzy Prószyn´ski2 1 Zak´lad Zoologii WSRP 08–110 Siedlce, Poland 2Muzeum i Instytut Zoologii PAN, ul. Wilcza 64, 00–950 Warszawa, Poland Summary The genus Euophrys from Britain and Central Europe (excluding the Mediterranean) is redefined. Of seventeen species analysed, only E. frontalis (Walckenaer, 1802) and E. herbigrada (Simon, 1871) are proposed to represent Euophrys (sensu stricto). Genus Pseudeuophrys is reinstated to include four European species. Six species are listed in Talavera. The relationships between the three genera and their distribution are discussed. The status of three species has still to be clarified. Introduction subject of informal discussion for years, but in the majority of papers Euophrys is still the only Euophrys is one of the largest and yet one of genus considered. the most poorly known genera in the Salticidae. Logunov (1992) was the first to review the Prószyn´ski (1990) and Platnick (1993) listed position of some Palaearctic species. He sug- over 130 nominal species from Europe, Asia, gested limiting the genus Euophrys to the Africa, the Americas, and the Pacific islands. frontalis species group and excluding In its present sense, however, the genus is a E. erratica, E. lanigera and E. obsoleta. mixture of many groups of unrelated species, Logunov also transferred E. aequipes (O. P.- frequently included on the basis of small size Cambridge, 1871), E. monticola Kulczyn´ski, and some convergent similarities in genitalic 1884 and E. -

Characterization of Farmington Virus, a Novel Virus from Birds That Is Distantly Related to Members of the Family Rhabdoviridae
Palacios et al. Virology Journal 2013, 10:219 http://www.virologyj.com/content/10/1/219 RESEARCH Open Access Characterization of Farmington virus, a novel virus from birds that is distantly related to members of the family Rhabdoviridae Gustavo Palacios1†, Naomi L Forrester2,3,4†, Nazir Savji5,7†, Amelia P A Travassos da Rosa2, Hilda Guzman2, Kelly DeToy5, Vsevolod L Popov2,4, Peter J Walker6, W Ian Lipkin5, Nikos Vasilakis2,3,4 and Robert B Tesh2,4* Abstract Background: Farmington virus (FARV) is a rhabdovirus that was isolated from a wild bird during an outbreak of epizootic eastern equine encephalitis on a pheasant farm in Connecticut, USA. Findings: Analysis of the nearly complete genome sequence of the prototype CT AN 114 strain indicates that it encodes the five canonical rhabdovirus structural proteins (N, P, M, G and L) with alternative ORFs (> 180 nt) in the N and G genes. Phenotypic and genetic characterization of FARV has confirmed that it is a novel rhabdovirus and probably represents a new species within the family Rhabdoviridae. Conclusions: In sum, our analysis indicates that FARV represents a new species within the family Rhabdoviridae. Keywords: Farmington virus (FARV), Family Rhabdoviridae, Next generation sequencing, Phylogeny Background Results Therhabdovirusesarealargeanddiversegroupofsingle- Growth characteristics stranded, negative sense RNA viruses that infect a wide Three litters of newborn (1–2 day old) ICR mice with aver- range of vertebrates, invertebrates and plants [1]. The family agesizeof10pupswereinoculated intracerebrally (ic) with Rhabdoviridae is currently divided into nine approved ge- 15–20 μl, intraperitoneally (ip) with 100 μlorsubcutane- nera (Vesiculovirus, Perhavirus, Ephemerovirus, Lyssavirus, ously (sc) with 100 μlofastockofVero-grownFARV(CT Tibrovirus, Sigmavirus, Nucleorhabdovirus, Cytorhabdovirus AN 114) virus containing approximately 107 plaque and Novirhabdovirus);however,alargenumberofanimal forming units (PFU) per ml. -
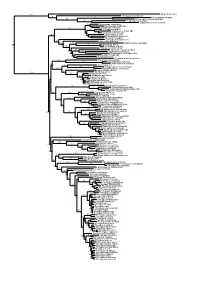
Figs1 ML Tree.Pdf
100 Megaderma lyra Rhinopoma hardwickei 71 100 Rhinolophus creaghi 100 Rhinolophus ferrumequinum 100 Hipposideros armiger Hipposideros commersoni 99 Megaerops ecaudatus 85 Megaerops niphanae 100 Megaerops kusnotoi 100 Cynopterus sphinx 98 Cynopterus horsfieldii 69 Cynopterus brachyotis 94 50 Ptenochirus minor 86 Ptenochirus wetmorei Ptenochirus jagori Dyacopterus spadiceus 99 Sphaerias blanfordi 99 97 Balionycteris maculata 100 Aethalops alecto 99 Aethalops aequalis Thoopterus nigrescens 97 Alionycteris paucidentata 33 99 Haplonycteris fischeri 29 Otopteropus cartilagonodus Latidens salimalii 43 88 Penthetor lucasi Chironax melanocephalus 90 Syconycteris australis 100 Macroglossus minimus 34 Macroglossus sobrinus 92 Boneia bidens 100 Harpyionycteris whiteheadi 69 Harpyionycteris celebensis Aproteles bulmerae 51 Dobsonia minor 100 100 80 Dobsonia inermis Dobsonia praedatrix 99 96 14 Dobsonia viridis Dobsonia peronii 47 Dobsonia pannietensis 56 Dobsonia moluccensis 29 Dobsonia anderseni 100 Scotonycteris zenkeri 100 Casinycteris ophiodon 87 Casinycteris campomaanensis Casinycteris argynnis 99 100 Eonycteris spelaea 100 Eonycteris major Eonycteris robusta 100 100 Rousettus amplexicaudatus 94 Rousettus spinalatus 99 Rousettus leschenaultii 100 Rousettus aegyptiacus 77 Rousettus madagascariensis 87 Rousettus obliviosus Stenonycteris lanosus 100 Megaloglossus woermanni 100 91 Megaloglossus azagnyi 22 Myonycteris angolensis 100 87 Myonycteris torquata 61 Myonycteris brachycephala 33 41 Myonycteris leptodon Myonycteris relicta 68 Plerotes anchietae