Diversity and Differentiation of in Bavaria
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mitteilungsblatt September 2020
Mitteilungsblatt für den Markt Pyrbaum mit amtlichen Bekanntmachungen Nr. 9 15. September 2020 44. Jahrgang Verabschiedung von Bürgermeister Guido Belzl und der ausgeschiedenen Markträtinnen und Markträte aus der Wahlperiode 2014-2020 (Foto Heike Regnet) Im Rahmen einer kleinen Abschiedsfeier wurde Bürgermeister Guido Belzl nach 18 Jahren Amtszeit verabschiedet. Ein besonderer Dank erging an die ausscheidenden Gemeinderätinnen und Gemeinderäte. Rekordwert von 4,05 Milliarden Euro an (Vorjahr: ~ 3,9 Mrd. €). Bekanntmachungen Der Verwaltungshaushalt des Marktes Pyrbaum wird auf der Einnah- menseite mit Schlüsselzuweisungen in Höhe von 816.000 € (Vorjahr: 1.128.196 €) verstärkt. Bekanntmachung zum Jahresabschluss Gleichzeitig steigt auf der Ausgabenseite der Anteil an der Kreisumlage um rund 608.000 €. Der Landkreis Neumarkt hat im Haushaltsjahr 2020 des Kommunalunternehmens den Umlagesatz auf einheitlich 36 % festgesetzt. Die Kreisumlagebelas- Markt Pyrbaum gem. Art. 90 ff. tung erhöht sich somit auf 2.471.000 € und belastet den Verwaltungs- Gemeindeordnung für den Freistaat haushalt damit bedeutender als im Jahr zuvor. Bayern in Verbindung mit § 27 Abs. 3 der Vermögenshaushalt Kommunalen Unternehmensverordnung Den größten Ausgabenblock bilden im Vermögenshaushalt die Baumaß- Für das Kommunalunternehmen Markt Pyrbaum, welches satzungsge- nahmen mit einem Volumen von 10.941.700 €, hier sind u.a. folgende mäß als Hauptaufgabe die Errichtung und die Vermietung der Pflegeein- Projekte enthalten (angegeben ist der jeweilige Kostenanteil für das richtung erfüllt, ist für das Jahr 2019 der Jahresabschluss bestehend aus Haushaltsjahr 2020): Bilanz, Gewinn- und Verlustrechnung unter Einbeziehung der Buchführung - Sanierungsmaßnahme „Neumarkter Straße“ 2.535.000 € und des Lageberichts von einem Sachverständigen geprüft worden. Vom - Sanierungsmaßnahme „Schulstraße“ 1.530.000 € beauftragten Prüfungsbüro sind keine Einwendungen erhoben worden. -

Infoblatt-Schierling-Langquaid.Pdf
KARTE FÜR ZOOM (mehr Details/Dünnere Linen/kleine Schrift) Schwandorf Teublitz 41 Maxhütte-Haidhof Burglengenfeld Grafenwinn Rohrbach ÖPNV-OffensiveKlein Ramspau Landkreise Regensburg und Kelheim Kallmünz Karlstein 15 Deutlich verbessertes107 Wochenendangebot auf der Linie 25 Langquaid – Schierling – Eggmühl Bhf Holzheim Diesenbach Heitzenhofen Regenstauf A3 SteinsbergGemeinsam verbessern die Landkreise Alternative zum Autoverkehr Falkenstein – 116 Wolfsegg Regensburg und Kelheim die Anbindung Cham von Langquaid142 und Schierling an den „Vorrangiges Ziel unserer ÖPNV-Offensive Duggendorf 14 Bahnhof Eggmühl. Das Projekt ist Teil der 2020 ist es, den öffentlichen Nahverkehr Regendorf 12 Kaulhausen ÖPNV-Offensive 2020 des Landkreises Bernhardswaldnoch hochwertiger, schneller und damit noch 17 34 Beratzhausen Rohrdorf Hainsacker Regensburg. bedarfsorientierter zu gestalten. So kann der Pielenhofen 142 13 Zeitlarn öffentliche Nahverkehr immer mehr zu einer Dettenhofen Die Aufwertung bringt zum einen zusätzliche echten Alternative zum Autoverkehr werden“, Rodau Lorenzen Wenzenbach Busverbindungen117 an Samstagen. Dabei sind betont die Regensburger Landrätin Tanja Pettendorf Pielmühle Hemau die Abfahrtszeiten am Bahnhof Eggmühl auf Schweiger. 28 die ZugverbindungenLappersdorf ab Regensburg Hbf mit Deuerling Ankunft in Eggmühl um 19:00 Uhr und 21:00 Ihr Kelheimer Amtskollege Martin Neumeyer Oppersdorf Galling- Nittendorf Uhr abgestimmt. Zumkofen anderen fährt an Sonn- ergänzt: „Die Bus-Zug-Kombination zwischen Kareth und Feiertagen erstmalig ein Buszubringer zum Langquaid und dem Bahnhof Eggmühl mit Um- Undorf Bahnhof Eggmühl. Das zweistündliche Fahrten- steigemöglichkeit in und aus Richtung Regens- Kneiting angebot zwischen 8:30 Uhr und 21:00 UhrBarbing burg oder München verbessertIllkofen das ÖPNV-An- schafft in beiden Fahrtrichtungen Anschlüsse gebot für33 viele Bürgerinnen und Bürger und an dieRegensburg Regional-Express-Verbindungen aus und leistet so einen wichtigen Beitrag zur Stärkung A3 nach Regensburg sowie München. -

Die Regionalökonomischen Auswirkungen Des Flughafens Memmingen Auf Den Tourismus
100 2019 Die regionalökonomischen Auswirkungen des Flughafens Memmingen auf den Tourismus Alfred Bauer, Florian Dorn, Luisa Dörr, Stefanie Gäbler, Manuela Krause, Martin Mosler, Christiaan Niemeijer, Horst Penzkofer und Niklas Potrafke ifo Zentrum für öffentliche Finanzen und politische Ökonomie Die regionalökonomischen Auswirkungen des Flughafens Memmingen auf den Tourismus Studie im Auftrag des Bayerischen Staatsministeriums für Wirtschaft, Landesentwicklung und Energie Autoren Prof. Dr. Alfred Bauer Martin Mosler Florian Dorn Christiaan Niemeijer Luisa Dörr Horst Penzkofer Stefanie Gäbler Prof. Dr. Niklas Potrafke Manuela Krause Die Autoren danken den Praktikantinnen Christina Dannhorn und Charlotte Grynberg und den Praktikanten Felix Michalik und Julian Milek für die wertvolle Unterstützung bei der Erstellung dieser Studie. April 2019 ifo Zentrum für öffentliche Finanzen und politische Ökonomie ifo Zentrum für öffentliche Finanzen und politische Ökonomie Bibliografische Information der Deutschen Nationalbibliothek Die Deutsche Nationalbibliothek verzeichnet diese Publikation in der Deutschen Nationalbibliografie; detaillierte bibliografische Daten sind im Internet über http://dnb.d-nb.de abrufbar. ISBN: 978-3-95942-063-1 Alle Rechte, insbesondere das der Übersetzung in fremde Sprachen, vorbehalten. Ohne ausdrückliche Genehmigung des Verlags ist es auch nicht gestattet, dieses Buch oder Teile daraus auf photomechanischem Wege (Photokopie, Mikrokopie) oder auf andere Art zu vervielfältigen. © ifo Institut, München 2019 Druck: ifo Institut, -

AG Amberg Neueintragungen
In ( ) gesetzte Angaben der Anschrift und des Geschäftszweiges erfolgen ohne Gewähr: AG Amberg Neueintragungen HRB 6982 13.04.2021 MSR-Recycling GmbH, Steinberg am See, An der Staatsstraße 2145, 92449 Steinberg am See. Gesellschaft mit beschränkter Haftung. Die Gesellschafterversammlung vom 31.03.2021 hat die Änderung des § 1 Abs. 2 (Sitz der Gesellschaft, bisher Regensburg, Amtsgericht Regensburg HRB 17053) der Satzung beschlossen. Geschäftsanschrift: An der Staatsstraße 2145, 92449 Steinberg am See. Gegenstand des Unternehmens: Rückgewinnung sortierter Wertstoffe, Handel mit Abfällen, Altmaterialien, Reststoffen, Schrott und Metallen, Gebäudeabbruch, Beratung im Abfall- und Recyclingbereich sowie Handel mit und Verleih von Baumaschinen im weitesten Sinne. Stammkapital: 25.000,00 EUR. Ist nur ein Geschäftsführer bestellt, so vertritt er die Gesellschaft allein. Sind mehrere Geschäftsführer bestellt, so wird die Gesellschaft durch zwei Geschäftsführer oder durch einen Geschäftsführer gemeinsam mit einem Prokuristen vertreten. Bestellt: Geschäftsführer: Sperl, Thomas, Tegernheim, *06.08.1985, einzelvertretungsberechtigt; mit der Befugnis, im Namen der Gesellschaft mit sich im eigenen Namen oder als Vertreter eines Dritten Rechtsgeschäfte abzuschließen. HRB 6983 14.04.2021 HiTES-H2 GmbH, Sulzbach-Rosenberg, An der Maxhütte 1, 92237 Sulzbach-Rosenberg. Gesellschaft mit beschränkter Haftung. Gesellschaftsvertrag vom 04.03.2021. Geschäftsanschrift: An der Maxhütte 1, 92237 Sulzbach-Rosenberg. Gegenstand des Unternehmens: Erzeugung von Wasserstoff -

Frepzeptkarte LEGENDE
1 2 3 3 25 Frankenweg 3 3 24 5 21 F REIZEITKARTE 25 2 25 15 21 UNTERWEGS ZWISCHEN 15 F RÄNKISCHEM S EENLAND 15 14 Benediktinerabtei Plankstetten Burg Burgthann Berching Frankenweg U ND B AYERISCHEM J U RA 2 3 4 21 24 13 21 24 23 13 8 13 3 23 24 23 24 24 14 Burgfest in Burgthann Christoph Willibald Gluck Landpartie Main-Donau-Kanal bei Berching 14 5 6 3 20 14 18 15 12 17 2 2 2 1718 Erholung am alten Kanal Spitalstadl Freystadt Christoph Willibald Gluck Museum 22 7 8 22 22 Frankenweg 2 www.bayerischerjura.de www.fraenkisches-seenland.de 9 2 6 21 6 EGENDE 10 1 L 10 22 17 10 2 9 6 1 Überregionale Radwege 17 4-Sattel-Feste Steinbruch Winnberg Kreislehrgarten in Burgthann 2 9 10 Fränkischer Seenlandweg 10 Oberbuchfeld 1 11 Fünf-Flüsse-Radweg 2 10 5 1 7 Land der Zeugenberge Tour de Baroque Unterbuchfeld Radwegenetz der Landkreise 1 Frankenweg 23 9 12 1 12 2 7 17 19 7 Überregionale Wanderwege 2 11 19 2 7 Nothelferkapelle Möninger Berg 8 8 1 Allersberg-Hilpoltstein-Roth-Weg 19 10 1 Berchinger Weg 12 19 2 Deininger Weg 14 18 3 Der Seenländer 13 11 12 Eppeleinsweg 1 2 Frankenweg Frankenweg 6 17 2 Jurasteig Kulturwanderweg Mit AOM-Radtouren die Region kennenlern 1 10 König-Ludwig-Schlaufe (Jurasteig) 12 Mariahilf-Schlaufe (Jurasteig) 12 2 3 Schwarzachtalweg 20 Spange Jurasteig-Seenland 24 16 20 17 Wasser- und Mühlenweg 20 12 Zeugenbergrunde Örtliche Wanderwege Alte Knabenschule Berngau 4 1 An der Kleinen Roth 11 19 2 Auf der Wasserscheide 3 Bahnweg 11 Frankenweg 4 Benediktusweg 11 5 Berngauer Buchbergweg 12 6 Berngauer Nordschlaufe 7 Berngauer Südschlaufe -

Bürgerinformationsbroschüre
Leben & Wohlfühlen im Markt Pyrbaum BÜRGERINFORMATIONEN www.pyrbaum.de Egal ob Automobil- oder Pharmakonzerne, Vereine oder der Maler bzw.Egal Friseur ob Automobil- um’s Eck oder- Service, Pharmakonzerne, Qualität und Vereine Flexibilität oder gelten der Maler für alle -Egalbzw. und ob Friseur das Automobil- zu um’s einem Eck oder fairen - Service, Pharmakonzerne, Preis-Leistungs-Verhältnis. Qualität und Vereine Flexibilität oder dergelten Maler für bzw. Friseuralle - und um‘s das Eck zu – einem Service, fairen Qualität Preis-Leistungs-Verhältnis. und Flexibilität gelten für alle – Egal ob Automobil- oder Pharmakonzerne, Vereine oder der Maler Zumund umfangreichen das zu einem Angebotfairen Preis-Leistungs-Verhältnis. von dkpromotion gehören innovative Zum umfangreichenbzw. AngebotFriseur um’s von dkpromotionEck - Service, gehören Qualität innovative und Flexibilität gelten für Artikel aller Art u.a. Kugelschreiber, Feuerzeuge, USB Sticks, ZumArtikel umfangreichen aller Art u.a.alle Kugelschreiber, -Angebot und das vonzu einem dk Feuerzeuge, promotion fairen Preis-Leistungs-Verhältnis. gehören USB Sticks, innovative Mit 20-jähriger Kalender, Tassen, Caps, Textilien,... Mit 20-jähriger ArtikelKalender, aller Tassen, Art u. a. Caps, Kugelschreiber, Textilien,... Feuerzeuge, USB Sticks, Kalender, Berufserfahrung ... überraschend anders! Berufserfahrung Tassen, Caps, TextilienZum umfangreichen ... Angebot von dkpromotion gehören innovative gründete Dieter gründete Dieter AlleinAllein in ihrem imin ihrem Online-Shop Online OnlineArtikel Shop Shop sindaller sind mehr Artsind mehr u.a. mehrals Kugelschreiber,75.000 als als 75.000 75.000 Artikel Artikel Artikel verfügbar. Feuerzeuge, verfügbar. USB Sticks, Probst 2008 Mit 20-jähriger fon +49 (0) 9176 9950-15 Probst 2008 Mit 20-jähriger Kalender, Tassen, Caps, Textilien,... die Firma Berufserfahrung die Firma Berufserfahrung WWerbeartikelerbeartikel fax +49 (0) 9176 9950-16dkpromotion. -
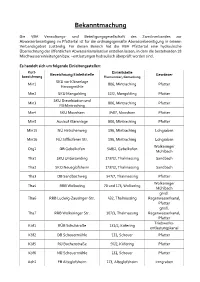
Bekanntmachung
Bekanntmachung Die VBA Verwaltungs- und Beteiligungsgesellschaft des Zweckverbandes zur Abwasserbeseitigung im Pfattertal ist für die ordnungsgemäße Abwasserbeseitigung in seinem Verbandsgebiet zuständig. Für diesen Bereich hat die VBA Pfattertal eine hydraulische Überrechnung der öffentlichen Abwasserkanalisation erstellen lassen, in dem die bestehenden 18 Mischwassereinleitungen bzw. –entlastungen hydraulisch überprüft worden sind. Es handelt sich um folgende Einleitungsstellen: Kurz- Einleitstelle Bezeichnung Einleitstelle Gewässer bezeichnung Flurnummer, Gemarkung SKU vor Kläranlage Min1 806, Mintraching Pfatter Herzogmühle Min2 SKU Mangolding 52/2, Mangolding Pfatter SKU Dieselstation und Min3 806, Mintraching Pfatter FB Mintraching Min4 SKU Moosham 3407, Moosham Pfatter Min5 Auslauf Kläranlage 806, Mintraching Pfatter Min15 NÜ Hetschenweg 196, Mintraching Lohgraben Min16 NÜ Siffkofener Str. 196, Mintraching Lohgraben Wolkeringer Otg1 DB Gebelkofen 548/2, Gebelkofen Mühlbach Tha1 SKU Untersanding 1737/2, Thalmassing Sandbach Tha2 SKO Neueglofsheim 1737/2, Thalmassing Sandbach Tha3 DB Sandbachweg 547/7, Thalmassing Pfatter Wolkeringer Tha4 RRB Wolkering 70 und 173, Wolkering Mühlbach gmdl. Tha6 RRB Ludwig-Zausinger-Str. 432, Thalmassing Regenwasserkanal, Pfatter gmdl. Tha7 RRB Wolkeringer Str. 107/3, Thalmassing Regenwasserkanal, Pfatter Triebwerks- Köf1 RÜB Schulstraße 131/2, Köfering entlastungskanal Köf2 DB Scheuermühle 131, Scheuer Pfatter Köf5 RÜ Buchenstraße 97/2, Köfering Pfatter Köf6 NÜ Scheuermühle 131, Scheuer Pfatter Aeh1 -

Glascontainerstandorte (PDF)
Liste der Stellplätze mit Glascontainern im Landkreis Neumarkt, Stand 02/2021 Sollte ein Stellplatz nicht mehr bestehen, bitten wir Sie um Mitteilung an folgende E-Mailadresse: [email protected] Ort Standplatz Berching Bahnhofstr. Berching Bauhof, Maria-Hilf-Str. (Wertstoffhof) Berching Burggriesbacher Str. Berching Post, Schulstr. Berching Erasbach, Franz-Schubert-Str. Berching Fribertshofen, Ortsstraße Berching Holnstein, Rudersdorfer Str. Berching Oening, Straße A Berching Plankstetten Berching Pollanten, Hauptstr. Berching Rappersdorf, Johannesstr. Berching Rudertshofen, Jugendheim Berching Sollngriesbach, Halberauweg Berching Staufersbuch, Beim Jugendheim Berching Thann, Ortsstraße Berching Wallnsdorf, Feuerwehrhaus Berching Weidenwang, Straße K Berg Hausheimer Str. Berg Waller Str. Berg Schulstr. Berg Gnadenberg, Gnadenberger Str. Ortsausgang Richtung Schleifmühle Berg Hausheim, Peter-und-Paul-Straße/Kaltenbachstraße Berg Kettenbach, Deponie (Wertstoffhof) Berg Loderbach, Loderbacher Hauptstraße, nahe der Brücke über die Schwarzach Berg Oberölsbach, Kreisstr. Richtung Sindlbach links nach Autobahnunterführung Berg Richtheim, Hölzlweg Berg Sindlbach, Hauptstr. Berg Stöckelsberg, Stöckelsberger Hauptstraße bei Hs.-Nr.36 Berg Unterölsbach, Barstenweg Berg Unterrohrenstadt, Am Schwall Berngau Pavelsbacher Str. Berngau Weiherstr. Berngau Röckersbühl, bei Trocknungsanlage. Berngau Tyrolsberg, Berngauer Straße Breitenbrunn Dürner Str. Breitenbrunn Badstr. Breitenbrunn Dürn, Ortsstraße Breitenbrunn Gimpertshausen, Dorfstraße -

210510 Mitteilungsblatt Mai 2021.Pdf
MITTEILUNGSBLATT für die Verwaltungsgemeinschaft Kallmünz www.vg-kallmuenz.de Mitgliedsgemeinden: Gemeinde Duggendorf Markt Kallmünz Gemeinde Holzheim a. Forst www.duggendorf.de www.kallmuenz.de www.holzheim-a-forst.de Geschäftsstelle der Verwaltungsgemeinschaft Kallmünz, Keltenweg 1, 93183 Kallmünz • Telefon (09473) 9401- 0 Telefax (09473) 9401-19 e-mail: [email protected] Öffnungszeiten: vormittags Montag, Dienstag, Donnerstag und Freitag von 8.00–12.00 Uhr nachmittags Dienstag von 13.30–17.00 Uhr, Donnerstag von 13.30 –18.00 Uhr ab sofort Mittwoch ganztägig geschlossen Öffnungszeiten der Wertstoffhöfe: Kallmünz Duggendorf Holzheim a. Forst Mittwoch von 17.00 bis 19.00 Uhr Freitag von 14.00 bis 16.30 Uhr Freitag von 14.30 bis 16.30 Uhr Freitag von 12.30 bis 16.30 Uhr Samstag von 9.30 bis 12.00 Uhr Samstag von 10.00 bis 12.00 Uhr Samstag von 9.00 bis 13.00 Uhr von Mai bis einschl. Oktober von Mai bis einschl. September Dienstag von 18.00 bis 19.00 Uhr Dienstag von 17.00 bis 19.00 Uhr nur Grüngutanlieferungen Öffnungszeiten der Gemeindebücherei Kallmünz jeden Dienstag von 16.00 bis 19.30 Uhr, Mittwochsausleihe siehe Aushang Bücherei 7.45–12.15 Uhr, Donnerstag 16.30–18.30 Uhr, Ferienzeiten nur donnerstags geöffnet. 42. Jahrgang Mai 2021 Nr. 5 Verwaltungsgemeinschaft Kallmünz Die Geschäftsstelle der Verwaltungsgemeinschaft Kallmünz ist am Freitag, 14.05.2021 und am Freitag, 04.06.2021 ganztägig geschlossen. Am 26. September 2021 findet die Bundestagswahl statt. Die Verwaltungsgemeinschaft Kallmünz sucht hierzu freiwillige, ehrenamtliche Wahlhelferinnen und Wahlhelfer. Bei uns bewerben können sich alle Volljährigen, mit deutscher Staatsangehörigkeit und Wohnsitz im Bereich der Verwaltungs- gemeinschaft Kallmünz. -

Heft 50 / April 2015 Schutzpreis Für Nichtmitlieder 2,00 € BJV Heft 50 2015 25.03.15 08:10 Seite 2 BJV Heft 50 2015 25.03.15 08:10 Seite 3
BJV_Heft_50_2015 25.03.15 08:10 Seite 1 JAGD UND NATURSCHUTZ Anerkannter Naturschutzverband LANDESJAGDVERBAND BAYERN e.V. BEZIRKSVERBAND REGENSBURG e.V. 50. Jubiläumsausgabe Einladung zur Ordentlichen Mitgliederversammlung 2015 Termine unter: www.jagd-regensburg.de JAHRGANG 25 Heft 50 / April 2015 Schutzpreis für Nichtmitlieder 2,00 € BJV_Heft_50_2015 25.03.15 08:10 Seite 2 BJV_Heft_50_2015 25.03.15 08:10 Seite 3 Inhalt Einladung zur Hubertusmesse 2015 . 4 Aus der Redaktion Im Visier . 5 Liebe Jägerinnen, liebe Jäger, Hubertus Mühlig, 1. Vorsitzender über fast ein Jahrzehnt hat Dr. Otto Spanner in der „Jagd und Naturschutz“ zu Themen aus Jagd und Naturschutz berichtet. Es fing an unter der Überschrift „Die Seltenen“ mit der Vorstellung Schießsportzentrum Bockenberg . 6 seltener Blumen in unserem Land, weil er der vermutlich richti- gen Ansicht war, dass Jäger auch über Blumen mehr Bescheid Untere Jagdbehörde . 7–10 wissen sollten als nur über das Gänseblümchen oder den Lö - Karl Frank wen zahn. Aus diesen Anfängen heraus sind dann viele Berichte über seine Erlebnisse bei der Jagd entstanden, ergänzt durch die Jagderlebnisse von Manfred H. J. Gold, die unsere Zeitschrift, UVV Jagd . 11 weit über ein Organ der Informationsvermittlung, zu einem von der ersten bis zur letzten Seite lesenswerten Heft aufgewertet Hygieneverordnung Tierische Lebensmittel . 12 haben. Otto Spanner hat mit dem Beitrag im letzten Heft vorerst seinen letzten Artikel geliefert. Es bleibt offen, ob er sich in die- Hundewesen in der BJV Kreisgruppe . 13 sem Heft noch einmal zu Wort melden wird. Den Jägerinnen und Jägern wird er in dankbarer Erinnerung bleiben. Klaus Wiesner Ihr Kai Taeger Hubertusmesse in der Alten Kapelle 2014 . -

Bildungsregion Im Landkreis Berchtesgadener Land
BEWERBUNGSKONZEPT BildunGsregion im Landkreis Berchtesgadener Land 1 Impressum Herausgeber Landkreis Berchtesgadener Land Salzburger Straße 64 83435 Bad Reichenhall www.lra-bgl.de Bearbeitung – redaktionelle Verantwortung Landratsamt Berchtesgadener Land Büro des Landrats (Bearbeiter: Stefan Neiber) Dank Wir danken herzlich den vielen regionalen Akteuren, die mit hohem persönlichem Einsatz und fundiertem Sachverstand den Ergebnisbericht erarbeitet haben, für ihr großes Engagement sowie den überregionalen Ansprechpartnern und Koordinatoren für ihre stete Unterstützung. Sonstige Hinweise Wir haben alle aufgeführten Daten, Informationen und Darstellungen nach bestem Wissen und Gewissen erarbeitet und geprüft. Es kann aber keine Gewähr für deren Aktualität, Richtigkeit und Vollständigkeit übernommen werden. Datum 17. Juni 2016 2 Inhaltsverzeichnis Kapitel Detailbezeichnung Seite Vorwort Landrat Georg Grabner 4 Kapitel 1 Der Weg zur Bildungsregion 5-14 1.1 Vorberatungen 5 1.2 Erstes Dialogforum am 27. Mai 2014 5 1.3 Arbeitsphase von Juni 2014 bis April 2015 6 1.4 Zweites Dialogforum am 1. Oktober 2015 11 Kapitel 2 Zusammenfassung der Bewerbung 15-28 2.1 Ausgangssituation im Berchtesgadener Land 15 2.2 Ergebnisse in den Handlungsfeldern 20 2.3 Maßnahmenplan ab 2016 – Grundsätze der Umsetzungsstrategie 26 Kapitel 3 Gesamtkonzept der Bewerbung 29-90 Kapitel 3.1 Übergänge organisieren und begleiten 29-41 3.1.1 Übergang Kindergarten – Grundschule 29 3.1.2 Übergang Grundschule – Gymnasium 32 3.1.3 Übergang Grundschule – Mittelschule/Realschule 35 3.1.4 Übergang Schule – Berufliche Bildung – Studium 37 3.1.5 Übergänge für Schüler mit sonderpädagogischem Förderbedarf 40 Kapitel 3.2 Schulische und außerschulische Bildungsangebote und Bildungsträger 42-63 vernetzen – Schulen in die Region öffnen 3.2.1 Verbesserung der Bedingungen und Arbeitsweisen von Schulen 42 3.2.2 Vernetzung aller Bildungsanbieter 47 3.2.3 Erwachsenenbildung/Lebenslanges Lernen 51 3.2.4 Bildungsnetz bzw. -

Selected Cluster Successes
Selected Cluster Successes Results from the promotion of innovative services Imprint The Federal Ministry for Economic Affairs and Energy has been awarded the berufundfamilie® Published by audit certificate for its family-friendly HR policy. Federal Ministry for Economic Affairs and Energy (BMWi) The certificate is awarded by berufundfamilie Public Relations Division gGmbH, an initiative of the non-profit Hertie 11019 Berlin, Germany Foundation. www.bmwi.de Text and editing Participating clusters VDI/VDE Innovation + Technik GmbH Design and production VDI/VDE Innovation + Technik GmbH, AZ As of March 2015 Picture credits © ARochau / fotolia.com (cover) This brochure is published as part of the public This and other brochures are available from: relations work of the Federal Ministry for Economic Federal Ministry for Economic Affairs and Energy Affairs and Energy. It is distributed free of charge Public Relations Division and is not intended for sale. The distribution of this E-mail: [email protected] brochure at campaign events or at information stands www.bmwi.de run by political parties is prohibited, and political Order service: party-related information or advertising shall not be Phone: +49 30 182722721 inserted into, printed on, or affixed to this publication. Fax: +49 30 18102722721 3 Contents 1. Foreword .....................................................................................................................................................................................................................................