Molekulare Phylogenie Der Pteriomorphen Bivalvia (Mollusca)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

BIOPAPUA Expedition Highlighting Deep-Sea Benthic Biodiversity of Papua New- Guinea
Biopapua Expedition – Progress report MUSÉUM NATIONAL D'HISTOIRE NATURELLE 57 rue Cuvier 75005 PARIS‐ France BIOPAPUA Expedition Highlighting deep-sea benthic Biodiversity of Papua New- Guinea Submitted by: Muséum National d'Histoire Naturelle (MNHN) Represented by (co‐PI): Dr Sarah Samadi (Researcher, IRD) Dr Philippe Bouchet (Professor, MNHN) Dr Laure Corbari (Research associate, MNHN) 1 Biopapua Expedition – Progress report Contents Foreword 3 1‐ Our understanding of deep‐sea biodiversity of PNG 4 2 ‐ Tropical Deep‐Sea Benthos program 5 3‐ Biopapua Expedition 7 4‐ Collection management 15 5‐ Preliminary results 17 6‐ Outreach and publications 23 7‐ Appendices 26 Appendix 1 27 NRI, note n°. 302/2010 on 26th march, 2010, acceptance of Biopapua reseach programme Appendix 2 28 Biopapua cruise Report, submitted by Ralph MANA (UPNG) A Report Submitted to School of Natural and Physical Sciences, University of Papua New Guinea Appendix 3 39 Chan, T.Y (2012) A new genus of deep‐sea solenocerid shrimp (Crustacea: Decapoda: Penaeoidea) from the Papua New Guinea. Journal of Crustacean Biology, 32(3), 489‐495. Appendix 4 47 Pante E, Corbari L., Thubaut J., Chan TY, Mana R., Boisselier MC, Bouchet P., Samadi S. (In Press). Exploration of the deep‐sea fauna of Papua New Guinea. Oceanography Appendix 5 60 Richer de Forges B. & Corbari L. (2012) A new species of Oxypleurodon Miers, 1886 (Crustacea Brachyura, Majoidea) from the Bismark Sea, Papua New Guinea. Zootaxa. 3320: 56–60 Appendix 6 66 Taxonomic list: Specimens in MNHN and Taiwan collections 2 Biopapua Expedition – Progress report Foreword Biopapua cruise was a MNHN/IRD deep‐sea cruise in partnership with the School of Natural and Physical Sciences, University of Papua New Guinea. -

Marine Bivalve Molluscs
Marine Bivalve Molluscs Marine Bivalve Molluscs Second Edition Elizabeth Gosling This edition first published 2015 © 2015 by John Wiley & Sons, Ltd First edition published 2003 © Fishing News Books, a division of Blackwell Publishing Registered Office John Wiley & Sons, Ltd, The Atrium, Southern Gate, Chichester, West Sussex, PO19 8SQ, UK Editorial Offices 9600 Garsington Road, Oxford, OX4 2DQ, UK The Atrium, Southern Gate, Chichester, West Sussex, PO19 8SQ, UK 111 River Street, Hoboken, NJ 07030‐5774, USA For details of our global editorial offices, for customer services and for information about how to apply for permission to reuse the copyright material in this book please see our website at www.wiley.com/wiley‐blackwell. The right of the author to be identified as the author of this work has been asserted in accordance with the UK Copyright, Designs and Patents Act 1988. All rights reserved. No part of this publication may be reproduced, stored in a retrieval system, or transmitted, in any form or by any means, electronic, mechanical, photocopying, recording or otherwise, except as permitted by the UK Copyright, Designs and Patents Act 1988, without the prior permission of the publisher. Designations used by companies to distinguish their products are often claimed as trademarks. All brand names and product names used in this book are trade names, service marks, trademarks or registered trademarks of their respective owners. The publisher is not associated with any product or vendor mentioned in this book. Limit of Liability/Disclaimer of Warranty: While the publisher and author(s) have used their best efforts in preparing this book, they make no representations or warranties with respect to the accuracy or completeness of the contents of this book and specifically disclaim any implied warranties of merchantability or fitness for a particular purpose. -
Field Identification Guide to the Living Marine Resources In
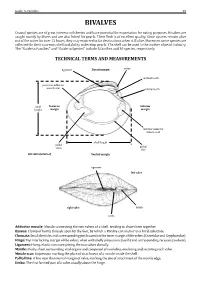
Guide to Families 29 BIVALVES Coastal species are of great interest to fisheries and have potential for exportation for eating purposes. Bivalves are caught mainly by divers and are also fished for pearls. Their flesh is of excellent quality. Since oysters remain alive out of the water for over 12 hours, they may exported to far destinations when still alive. Moreover, some species are collected for their nacreous shell and ability to develop pearls. The shell can be used in the mother of pearl industry. The “Guide to Families’’ andTECHNICAL ‘‘Guide to Species’’ TERMS include 5AND families MEASUREMENTS and 10 species, respectively. ligament Dorsal margin umbo posterior adductor cardinal tooth muscle scar lateral tooth Posterior Anterior margin margin shell height anterior adductor muscle scar pallial sinus pallial shell length line left valve (interior) Ventral margin ligament left valve right valve lunule umbo Adductor muscle: Byssus: Chomata: Muscle connecting the two valves of a shell, tending to draw them together. Hinge: Clump of horny threads spun by the foot, by which a Bivalve can anchor to a hard substrate. Ligament: Small denticles and corresponding pits located on the inner margin of the valves (Ostreidae and Gryphaeidae). Mantle: Top interlocking margin of the valves, often with shelly projections (teeth) and corresponding recesses (sockets). Muscle scar: Horny, elastic structure joining the two valves dorsally. Pallial line: Fleshy sheet surrounding vital organs and composed of two lobes, one lining and secreting each valve. Umbo: Impression marking the place of attachment of a muscle inside the shell. A line near the internal margin of valve, marking the site of attachment of the mantle edge. -

Diversity of Malacofauna from the Paleru and Moosy Backwaters Of
Journal of Entomology and Zoology Studies 2017; 5(4): 881-887 E-ISSN: 2320-7078 P-ISSN: 2349-6800 JEZS 2017; 5(4): 881-887 Diversity of Malacofauna from the Paleru and © 2017 JEZS Moosy backwaters of Prakasam district, Received: 22-05-2017 Accepted: 23-06-2017 Andhra Pradesh, India Darwin Ch. Department of Zoology and Aquaculture, Acharya Darwin Ch. and P Padmavathi Nagarjuna University Nagarjuna Nagar, Abstract Andhra Pradesh, India Among the various groups represented in the macrobenthic fauna of the Bay of Bengal at Prakasam P Padmavathi district, Andhra Pradesh, India, molluscs were the dominant group. Molluscs were exploited for Department of Zoology and industrial, edible and ornamental purposes and their extensive use has been reported way back from time Aquaculture, Acharya immemorial. Hence the present study was focused to investigate the diversity of Molluscan fauna along Nagarjuna University the Paleru and Moosy backwaters of Prakasam district during 2016-17 as these backwaters are not so far Nagarjuna Nagar, explored for malacofauna. A total of 23 species of molluscs (16 species of gastropods belonging to 12 Andhra Pradesh, India families and 7 species of bivalves representing 5 families) have been reported in the present study. Among these, gastropods such as Umbonium vestiarium, Telescopium telescopium and Pirenella cingulata, and bivalves like Crassostrea madrasensis and Meretrix meretrix are found to be the most dominant species in these backwaters. Keywords: Malacofauna, diversity, gastropods, bivalves, backwaters 1. Introduction Molluscans are the second largest phylum next to Arthropoda with estimates of 80,000- 100,000 described species [1]. These animals are soft bodied and are extremely diversified in shape and colour. -

New Records of Three Deep-Sea Bathymodiolus Mussels (Bivalvia: Mytilida: Mytilidae) from Hydrothermal Vent and Cold Seeps in Taiwan
352 Journal of Marine Science and Technology, Vol. 27, No. 4, pp. 352-358 (2019) DOI: 10.6119/JMST.201908_27(4).0006 NEW RECORDS OF THREE DEEP-SEA BATHYMODIOLUS MUSSELS (BIVALVIA: MYTILIDA: MYTILIDAE) FROM HYDROTHERMAL VENT AND COLD SEEPS IN TAIWAN Meng-Ying Kuo1, Dun- Ru Kang1, Chih-Hsien Chang2, Chia-Hsien Chao1, Chau-Chang Wang3, Hsin-Hung Chen3, Chih-Chieh Su4, Hsuan-Wien Chen5, Mei-Chin Lai6, Saulwood Lin4, and Li-Lian Liu1 Key words: new record, Bathymodiolus, deep-sea, hydrothermal vent, taiwanesis (von Cosel, 2008) is the only reported species of cold seep, Taiwan. this genus from Taiwan. It was collected from hydrothermal vents near Kueishan Islet off the northeast coast of Taiwan at depths of 200-355 m. ABSTRACT Along with traditional morphological classification, mo- The deep sea mussel genus, Bathymodiolus Kenk & Wilson, lecular techniques are commonly used to study the taxonomy 1985, contains 31 species, worldwide. Of which, one endemic and phylogenetic relationships of deep sea mussels. Recently, species (Bathymodiolus taiwanesis) was reported from Taiwan the complete mitochondrial genomes have been sequenced (MolluscaBase, 2018). Herein, based on the mitochondrial COI from mussels of Bathymodiolus japonicus, B. platifrons and results, we present 3 new records of the Bathymodiolus species B. septemdierum (Ozawa et al., 2017). Even more, the whole from Taiwan, namely Bathymodiolus platifrons, Bathymodiolus genome of B. platifrons was reported with sequence length of securiformis, and Sissano Bathymodiolus sp.1 which were collected 1.64 Gb nucleotides (Sun et al., 2017). from vent or seep environments at depth ranges of 1080-1380 Since 2013, under the Phase II National energy program of m. -

Molluscs (Mollusca: Gastropoda, Bivalvia, Polyplacophora)
Gulf of Mexico Science Volume 34 Article 4 Number 1 Number 1/2 (Combined Issue) 2018 Molluscs (Mollusca: Gastropoda, Bivalvia, Polyplacophora) of Laguna Madre, Tamaulipas, Mexico: Spatial and Temporal Distribution Martha Reguero Universidad Nacional Autónoma de México Andrea Raz-Guzmán Universidad Nacional Autónoma de México DOI: 10.18785/goms.3401.04 Follow this and additional works at: https://aquila.usm.edu/goms Recommended Citation Reguero, M. and A. Raz-Guzmán. 2018. Molluscs (Mollusca: Gastropoda, Bivalvia, Polyplacophora) of Laguna Madre, Tamaulipas, Mexico: Spatial and Temporal Distribution. Gulf of Mexico Science 34 (1). Retrieved from https://aquila.usm.edu/goms/vol34/iss1/4 This Article is brought to you for free and open access by The Aquila Digital Community. It has been accepted for inclusion in Gulf of Mexico Science by an authorized editor of The Aquila Digital Community. For more information, please contact [email protected]. Reguero and Raz-Guzmán: Molluscs (Mollusca: Gastropoda, Bivalvia, Polyplacophora) of Lagu Gulf of Mexico Science, 2018(1), pp. 32–55 Molluscs (Mollusca: Gastropoda, Bivalvia, Polyplacophora) of Laguna Madre, Tamaulipas, Mexico: Spatial and Temporal Distribution MARTHA REGUERO AND ANDREA RAZ-GUZMA´ N Molluscs were collected in Laguna Madre from seagrass beds, macroalgae, and bare substrates with a Renfro beam net and an otter trawl. The species list includes 96 species and 48 families. Six species are dominant (Bittiolum varium, Costoanachis semiplicata, Brachidontes exustus, Crassostrea virginica, Chione cancellata, and Mulinia lateralis) and 25 are commercially important (e.g., Strombus alatus, Busycoarctum coarctatum, Triplofusus giganteus, Anadara transversa, Noetia ponderosa, Brachidontes exustus, Crassostrea virginica, Argopecten irradians, Argopecten gibbus, Chione cancellata, Mercenaria campechiensis, and Rangia flexuosa). -

The Contrasted Evolutionary Fates of Deep-Sea
The contrasted evolutionary fates of deep-sea chemosynthetic mussels (Bivalvia, Bathymodiolinae) Justine Thubaut, Nicolas Puillandre, Jean-Baptiste Faure, Corinne Cruaud, Sarah Samadi To cite this version: Justine Thubaut, Nicolas Puillandre, Jean-Baptiste Faure, Corinne Cruaud, Sarah Samadi. The con- trasted evolutionary fates of deep-sea chemosynthetic mussels (Bivalvia, Bathymodiolinae). Ecology and Evolution, Wiley Open Access, 2013, 3 (14), pp.4748-4766. 10.1002/ece3.749. hal-01250918 HAL Id: hal-01250918 https://hal.archives-ouvertes.fr/hal-01250918 Submitted on 11 Jan 2016 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License The contrasted evolutionary fates of deep-sea chemosynthetic mussels (Bivalvia, Bathymodiolinae)a Justine Thubaut1, Nicolas Puillandre1, Baptiste Faure2,3, Corinne Cruaud4 & Sarah Samadi1 1Departement Systematique et Evolution, Museum National d’Histoire Naturelle, Unite Mixte de Recherche 7138 (UPMC-IRD-MNHN-CNRS), “Systematique Adaptation et Evolution”, 75005 Paris, France 2Station Biologique de Roscoff, Unite Mixte de Recherche 7127, Centre National de la Recherche Scientifique, Universite Pierre et Marie Curie, 29680 Roscoff, France 3Biotope, Service Recherche et Developpement, BP58 34140 Meze, France 4Genoscope, CP 5706, 91057 Evry, France Keywords Abstract Bathymodiolinae, chemosynthetic ecosystem, deep-sea, evolution. -

Discovery of Chemosynthesis-Based Association on the Cretaceous Basal Leatherback Sea Turtle from Japan
Editors' choice Discovery of chemosynthesis-based association on the Cretaceous basal leatherback sea turtle from Japan ROBERT G. JENKINS, ANDRZEJ KAIM, KEI SATO, KAZUHIRO MORIYA, YOSHINORI HIKIDA, and REN HIRAYAMA Jenkins, R.G., Kaim, A., Sato, K., Moriya, K., Hikida, Y., and Hirayama, R. 2017. Discovery of chemosynthesis-based association on the Cretaceous basal leatherback sea turtle from Japan. Acta Palaeontologica Polonica 62 (4): 683–690. We report a Late Cretaceous chemosynthetic community fueled by decomposing basal leatherback sea turtle on the ocean floor in the western Pacific. The fossil association representing this community has been recovered from the matrix of a concretion containing a single carapace of Mesodermochelys sp. from Late Cretaceous outer shelf to upper slope deposit of northern Hokkaido, Japan. The carapace displays boreholes most likely performed by boring bivalves, and is associated with molluscan shells, mainly Provanna cf. nakagawensis and Thyasira tanabei. Since this association is similar to fauna already known from Late Cretaceous hydrocarbon seeps, sunken wood, and plesiosaur-falls in Hokkaido, it is suggested that all types of chemosynthesis-based communities in the Late Cretaceous of western Pacific may have belonged to the same regional pool of animals and were not yet fully differentiated into three independent types of com- munities as it is known today. This finding also indicates that the sulfophilic stage of the vertebrate-fall communities was supported not only by plesiosaur carcasses, which were previously reported, but also by sea turtle carcasses. It highlights the possibility of surviving vertebrate-fall communities through the end-Cretaceous mass extinction event on carcasses of sea turtles which are the only large marine vertebrates surviving this event. -

Abstract Volume
ABSTRACT VOLUME August 11-16, 2019 1 2 Table of Contents Pages Acknowledgements……………………………………………………………………………………………...1 Abstracts Symposia and Contributed talks……………………….……………………………………………3-225 Poster Presentations…………………………………………………………………………………226-291 3 Venom Evolution of West African Cone Snails (Gastropoda: Conidae) Samuel Abalde*1, Manuel J. Tenorio2, Carlos M. L. Afonso3, and Rafael Zardoya1 1Museo Nacional de Ciencias Naturales (MNCN-CSIC), Departamento de Biodiversidad y Biologia Evolutiva 2Universidad de Cadiz, Departamento CMIM y Química Inorgánica – Instituto de Biomoléculas (INBIO) 3Universidade do Algarve, Centre of Marine Sciences (CCMAR) Cone snails form one of the most diverse families of marine animals, including more than 900 species classified into almost ninety different (sub)genera. Conids are well known for being active predators on worms, fishes, and even other snails. Cones are venomous gastropods, meaning that they use a sophisticated cocktail of hundreds of toxins, named conotoxins, to subdue their prey. Although this venom has been studied for decades, most of the effort has been focused on Indo-Pacific species. Thus far, Atlantic species have received little attention despite recent radiations have led to a hotspot of diversity in West Africa, with high levels of endemic species. In fact, the Atlantic Chelyconus ermineus is thought to represent an adaptation to piscivory independent from the Indo-Pacific species and is, therefore, key to understanding the basis of this diet specialization. We studied the transcriptomes of the venom gland of three individuals of C. ermineus. The venom repertoire of this species included more than 300 conotoxin precursors, which could be ascribed to 33 known and 22 new (unassigned) protein superfamilies, respectively. Most abundant superfamilies were T, W, O1, M, O2, and Z, accounting for 57% of all detected diversity. -

Pacific Lithophaga (Bivalvia, Mytilidae) from Recent French Expeditions with the Description of Two New Species
Boll. Malacol., 48: 73-102 (2/2012) Pacific Lithophaga (Bivalvia, Mytilidae) from recent French expeditions with the description of two new species Karl Kleemann*() & Philippe Maestrati# *Institute Abstract of Palaeontology, Pacific specimens of Lithophaga and its subgenus Leiosolenus, collected during recent French expeditions Centre of Earth Sciences, to New Caledonia, Vanuatu, the Philippines and French Polynesia, were determined and described, inclu- University of Vienna, ding two new species, Lithophaga (Leiosolenus) paraplumula n. sp. and Lithophaga (Leiosolenus) subatte- Althanstr. 14, 1090 Wien, Austria, nuata n. sp. From the twenty species, three belong to Lithophaga s.s. and seventeen to the subgenus karl.kleemann@ Leiosolenus. In order to help identification of the two new species and some others, selected specimens univie.ac.at, are figured in left lateral, right lateral and dorsal view. A taxonomic key is provided for determination. () Corresponding author Key words: Boring bivalves, Leiosolenus, Lithophaga, taxonomy, new species, Pacific. #Muséum national d’Histoire naturelle de Paris, Département Riassunto de Systématique et [Specie pacifiche di Lithophaga (Bivalvia, Mytilidae) da recenti spedizioni francesi, con la descrizione di due Evolution, UMR 7138, nuove specie]. Il presente lavoro presenta lo studio di esemplari Lithophaga e del suo sottogenere Leioso- 55 rue Buffon, lenus, raccolti in Nuova Caledonia, Vanuatu, Filippine e nella Polinesia Francese, in occasione di spedizioni 75005 Paris, France, francesi. Tutti gli esemplari vengono identificati a livello specifico, e descritti. Vengono introdotte due philippe.maestrati@ nuove specie: Lithophaga (Leiosolenus) paraplumula n. sp. e Lithophaga (Leiosolenus) subattenuata n. sp. mnhn.fr Su venti specie esaminate, tre appartengono a Lithophaga s.s. e diciassette al sottogenere Leiosolenus. -

TREATISE ONLINE Number 48
TREATISE ONLINE Number 48 Part N, Revised, Volume 1, Chapter 31: Illustrated Glossary of the Bivalvia Joseph G. Carter, Peter J. Harries, Nikolaus Malchus, André F. Sartori, Laurie C. Anderson, Rüdiger Bieler, Arthur E. Bogan, Eugene V. Coan, John C. W. Cope, Simon M. Cragg, José R. García-March, Jørgen Hylleberg, Patricia Kelley, Karl Kleemann, Jiří Kříž, Christopher McRoberts, Paula M. Mikkelsen, John Pojeta, Jr., Peter W. Skelton, Ilya Tëmkin, Thomas Yancey, and Alexandra Zieritz 2012 Lawrence, Kansas, USA ISSN 2153-4012 (online) paleo.ku.edu/treatiseonline PART N, REVISED, VOLUME 1, CHAPTER 31: ILLUSTRATED GLOSSARY OF THE BIVALVIA JOSEPH G. CARTER,1 PETER J. HARRIES,2 NIKOLAUS MALCHUS,3 ANDRÉ F. SARTORI,4 LAURIE C. ANDERSON,5 RÜDIGER BIELER,6 ARTHUR E. BOGAN,7 EUGENE V. COAN,8 JOHN C. W. COPE,9 SIMON M. CRAgg,10 JOSÉ R. GARCÍA-MARCH,11 JØRGEN HYLLEBERG,12 PATRICIA KELLEY,13 KARL KLEEMAnn,14 JIřÍ KřÍž,15 CHRISTOPHER MCROBERTS,16 PAULA M. MIKKELSEN,17 JOHN POJETA, JR.,18 PETER W. SKELTON,19 ILYA TËMKIN,20 THOMAS YAncEY,21 and ALEXANDRA ZIERITZ22 [1University of North Carolina, Chapel Hill, USA, [email protected]; 2University of South Florida, Tampa, USA, [email protected], [email protected]; 3Institut Català de Paleontologia (ICP), Catalunya, Spain, [email protected], [email protected]; 4Field Museum of Natural History, Chicago, USA, [email protected]; 5South Dakota School of Mines and Technology, Rapid City, [email protected]; 6Field Museum of Natural History, Chicago, USA, [email protected]; 7North -

Download Full Article in PDF Format
Une nouvelle espèce de Bathymodiolinae (Mollusca, Bivalvia, Mytilidae) associée à des os de baleine coulés en Méditerranée Jacques PELORCE 289 voie Les Magnolias, F-30240 Le Grau du Roi (France) [email protected] Jean-Maurice POUTIERS Muséum national d’Histoire naturelle, Département Systématique et Évolution, case postale 51, 57 rue Cuvier, F-75231 Paris cedex 05 (France) [email protected] Pelorce J. & Poutiers J.-M. 2009. — Une nouvelle espèce de Bathymodiolinae (Mollusca, Bivalvia, Mytilidae) associée à des os de baleine coulés en Méditerranée. Zoosystema 31 (4) : 975-985. RÉSUMÉ Les espèces du genre Idas Jeff reys, 1876 (Mollusca, Bivalvia, Mytilidae), associées à des substrats organiques coulés en profondeur en Méditerranée et dans l’Atlantique nord-est, sont passées en revue et une nouvelle espèce associée à de vieux os de baleine coulés « Idas » cylindricus n. sp., incluse provisoirement MOTS CLÉS dans le genre Idas s.l., est décrite du Golfe du Lion (Méditerranée occidentale). Mollusca, Bivalvia, Elle est comparée avec Idas (s.l.) simpsoni (Marshall, 1900) qui est l’espèce la Mytilidae, plus proche, et avec les autres espèces voisines de l’Atlantique nord-est et de Bathymodiolinae, Méditerranée. La nouvelle espèce se caractérise par sa taille importante pour le Idas, Méditerranée, genre, sa forme renfl ée et son profi l rectangulaire, son ligament interne épais et nouvelle espèce. marron et la position très avancée de l’umbo. ABSTRACT A new species of bathymodioline mussel (Mollusca, Bivalvia, Mytilidae) associated with sunken whale bones in the Mediterranean Sea. Species of the genus Idas Jeff reys, 1876 (Mollusca, Bivalvia, Mytilidae), associated with sunken organic substrates in the Mediterranean Sea and the North-East Atlantic Ocean, are reviewed.