INTRASPECIFIC POLYMORPHISM of RIBOSOMAL DNA LOCI NUMBER and MORPHOLOGY in Brachypodium Pinnatum and Brachypodium Sylvaticum
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Notes on Identification Works and Difficult and Under-Recorded Taxa
Notes on identification works and difficult and under-recorded taxa P.A. Stroh, D.A. Pearman, F.J. Rumsey & K.J. Walker Contents Introduction 2 Identification works 3 Recording species, subspecies and hybrids for Atlas 2020 6 Notes on individual taxa 7 List of taxa 7 Widespread but under-recorded hybrids 31 Summary of recent name changes 33 Definition of Aggregates 39 1 Introduction The first edition of this guide (Preston, 1997) was based around the then newly published second edition of Stace (1997). Since then, a third edition (Stace, 2010) has been issued containing numerous taxonomic and nomenclatural changes as well as additions and exclusions to taxa listed in the second edition. Consequently, although the objective of this revised guide hast altered and much of the original text has been retained with only minor amendments, many new taxa have been included and there have been substantial alterations to the references listed. We are grateful to A.O. Chater and C.D. Preston for their comments on an earlier draft of these notes, and to the Biological Records Centre at the Centre for Ecology and Hydrology for organising and funding the printing of this booklet. PAS, DAP, FJR, KJW June 2015 Suggested citation: Stroh, P.A., Pearman, D.P., Rumsey, F.J & Walker, K.J. 2015. Notes on identification works and some difficult and under-recorded taxa. Botanical Society of Britain and Ireland, Bristol. Front cover: Euphrasia pseudokerneri © F.J. Rumsey. 2 Identification works The standard flora for the Atlas 2020 project is edition 3 of C.A. Stace's New Flora of the British Isles (Cambridge University Press, 2010), from now on simply referred to in this guide as Stae; all recorders are urged to obtain a copy of this, although we suspect that many will already have a well-thumbed volume. -

European Glacial Relict Snails and Plants: Environmental Context of Their Modern Refugial Occurrence in Southern Siberia
bs_bs_banner European glacial relict snails and plants: environmental context of their modern refugial occurrence in southern Siberia MICHAL HORSAK, MILAN CHYTRY, PETRA HAJKOV A, MICHAL HAJEK, JIRI DANIHELKA, VERONIKA HORSAKOV A, NIKOLAI ERMAKOV, DMITRY A. GERMAN, MARTIN KOCI, PAVEL LUSTYK, JEFFREY C. NEKOLA, ZDENKA PREISLEROVA AND MILAN VALACHOVIC Horsak, M., Chytry, M., Hajkov a, P., Hajek, M., Danihelka, J., Horsakov a,V.,Ermakov,N.,German,D.A.,Ko cı, M., Lustyk, P., Nekola, J. C., Preislerova, Z. & Valachovic, M. 2015 (October): European glacial relict snails and plants: environmental context of their modern refugial occurrence in southern Siberia. Boreas, Vol. 44, pp. 638–657. 10.1111/bor.12133. ISSN 0300-9483. Knowledge of present-day communities and ecosystems resembling those reconstructed from the fossil record can help improve our understanding of historical distribution patterns and species composition of past communities. Here, we use a unique data set of 570 plots explored for vascular plant and 315 for land-snail assemblages located along a 650-km-long transect running across a steep climatic gradient in the Russian Altai Mountains and their foothills in southern Siberia. We analysed climatic and habitat requirements of modern populations for eight land-snail and 16 vascular plant species that are considered characteristic of the full-glacial environment of central Europe based on (i) fossil evidence from loess deposits (snails) or (ii) refugial patterns of their modern distribu- tions (plants). The analysis yielded consistent predictions of the full-glacial central European climate derived from both snail and plant populations. We found that the distribution of these 24 species was limited to the areas with mean annual temperature varying from À6.7 to 3.4 °C (median À2.5 °C) and with total annual precipitation vary- ing from 137 to 593 mm (median 283 mm). -

Species-Rich Grassland and National Vegetation Classification Virtual Practical
EDUCATION RESOURCE Species-rich Grassland and National Vegetation Classification Virtual Practical This virtual species-richness assessment and NVC practical uses real images that were taken through June, July and August. These are a record of species’ presence in 1x1m quadrats, but not percentage cover. Your task is to identify the species to determine if this is species-rich grassland and to classify the NVC community. You need to: • Identify the species in each image using the list below and on the next page and identification guides, for example The Wild Flower Key by Francis Rose or New Flora of the British Isles (fourth edition) by Clive Stace. • Play the video presentation and pause to examine every picture in each quadrat. Images are taken to emphasise one species, but sometimes they contain more than one. You can record all the species that you can identify from each image. • Use the data collection form to show species presence in each quadrat by putting a tick or a ‘P’ for presence in each quadrat where a species was present. • To determine whether the grassland classifies as ‘species-rich), add up the species richness of each quadrat (excluding negative indicator species as listed in the recording form) and find the mean. • To find the NVC classification, use British Plant Communities Volume 3 Grasslands and Montane Communities (Ed. J Rodwell) and the keys to the vegetation of calcicolous and mestrophic grasslands. Assess which community this sample fits into. Do you agree with this assessment and why? • Use the MAVIS software to classify the habitat type. -

Global Relationships Between Plant Functional Traits and Environment in Grasslands
GLOBAL RELATIONSHIPS BETWEEN PLANT FUNCTIONAL TRAITS AND ENVIRONMENT IN GRASSLANDS EMMA JARDINE A thesis submitted in partial fulfilment of the requirements for the degree of Doctor of Philosophy The University of Sheffield Department of Animal and Plant Sciences Submission Date July 2017 ACKNOWLEDGMENTS First of all I am enormously thankful to Colin Osborne and Gavin Thomas for giving me the opportunity to undertake the research presented in this thesis. I really appreciate all their invaluable support, guidance and advice. They have helped me to grow in knowledge, skills and confidence and for this I am extremely grateful. I would like to thank the students and post docs in both the Osborne and Christin lab groups for their help, presentations and cake baking. In particular Marjorie Lundgren for teaching me to use the Licor, for insightful discussions and general support. Also Kimberly Simpson for all her firey contributions and Ruth Wade for her moral support and employment. Thanks goes to Dave Simpson, Maria Varontsova and Martin Xanthos for allowing me to work in the herbarium at the Royal Botanic Gardens Kew, for letting me destructively harvest from the specimens and taking me on a worldwide tour of grasses. I would also like to thank Caroline Lehman for her map, her useful comments and advice and also Elisabeth Forrestel and Gareth Hempson for their contributions. I would like to thank Brad Ripley for all of his help and time whilst I was in South Africa. Karmi Du Plessis and her family and Lavinia Perumal for their South African friendliness, warmth and generosity and also Sean Devonport for sharing all the much needed teas and dub. -

Does Seed Modification and Nitrogen Addition Affect Seed Germination of Pulsatilla Grandis?*
ENVIRONMENTAL SCIENCES DOES SEED MODIFICATION AND NITROGEN ADDITION AFFECT SEED GERMINATION OF PULSATILLA GRANDIS?* M. Bochenková1, P. Karlík2, M. Hejcman1, P. Jiras1 1 Czech University of Life Sciences Prague, Faculty of Environmental Sciences, Department of Ecology, Czech Republic 2 Czech University of Life Sciences Prague, Faculty of Forestry and Wood Sciences, Department of Forest Ecology, Czech Republic Pulsatilla grandis is an endangered species in the Czech Republic and is protected in whole Europe because the number of its populations is declining. One of the possible causes is the deposition of atmospheric nitrogen. In our research, we investigated how nitrogen concentrations and seed appendage removal directly affect the species’ seed germination.Seeds were allowed to –1 germinate under laboratory conditions in water solutions of NH4NO3 ranging in concentration from 0 to 4239 mg N l . They were able to germinate up to the concentration of 848 mg N l–1 even when covered with mycelium, which supports the idea that they can tolerate being strongly infected by fungi. We also found a significant positive effect of seed appendage removal on seed germination. Seeds without appendages germinated, on the average, with 11% greater probability, compared to seeds with appendages. We conclude that the germination of P. grandis is not directly affected by high N concentrations in rain wa- ter, which can range from 10 to 13 mg N l–1 near large cities. Surprisingly, low concentrations of N (up to 34 mg N l–1) might even slightly support the seed germination of P. grandis. The negative effect of N deposition on seeds is indirect and acts in conjunction with the absence of management at localities. -

(Poaceae) and Characterization
EVOLUTION AND DEVELOPMENT OF VEGETATIVE ARCHITECTURE: BROAD SCALE PATTERNS OF BRANCHING ACROSS THE GRASS FAMILY (POACEAE) AND CHARACTERIZATION OF ARCHITECTURAL DEVELOPMENT IN SETARIA VIRIDIS L. P. BEAUV. By MICHAEL P. MALAHY Bachelor of Science in Biology University of Central Oklahoma Edmond, Oklahoma 2006 Submitted to the Faculty of the Graduate College of the Oklahoma State University in partial fulfillment of the requirements for the Degree of MASTER OF SCIENCE July, 2012 EVOLUTION AND DEVELOPMENT OF VEGETATIVE ARCHITECTURE: BROAD SCALE PATTERNS OF BRANCHING ACROSS THE GRASS FAMILY (POACEAE) AND CHARACTERIZATION OF ARCHITECTURAL DEVELOPMENT IN WEEDY GREEN MILLET ( SETARIA VIRIDIS L. P. BEAUV.) Thesis Approved: Dr. Andrew Doust Thesis Adviser Dr. Mark Fishbein Dr. Linda Watson Dr. Sheryl A. Tucker Dean of the Graduate College I TABLE OF CONTENTS Chapter Page I. Evolutionary survey of vegetative branching across the grass family (poaceae) ... 1 Introduction ................................................................................................................... 1 Plant Architecture ........................................................................................................ 2 Vascular Plant Morphology ......................................................................................... 3 Grass Morphology ....................................................................................................... 4 Methods ....................................................................................................................... -

Biodiversity Consultations
Rebecca Shelton Northamptonshire Biodiversity Wardell Armstrong LLP Records Centre Sir Henry Doulton House C/O The Wildlife Trust Lings House Forge Lane Billing Lings Etruria Northamptonshire Stoke-on-Trent NN3 8BE ST1 5NN Tel: 01604 405285 Fax: 01604 784835 [email protected] October 12th 2006 Dear Rebecca, Re: Ecological data search, Corby Thank you for approaching the NBRC with this enquiry. All the information that you have requested is contained within this report. This includes a map of the search area, statutory and non-statutory site details and a list of protected and notable species records from your specified search area. For definitions of these sites please refer to the document at the end of this report. Statutory sites No statutory sites were found within your search area. Non-statutory sites The following non-statutory sites are located within your specified search area. These sites have been labelled on the accompanying maps. Brookfield Plantation County Wildlife Site Brookfield Plantation Cutting County Wildlife Site Corby Old Quarries County Wildlife Site Corby Tunnel Quarries County Wildlife Site Weldon Churchyard County Wildlife Site Weldon Marsh County Wildlife Site Weldon Mound County Wildlife Site Weldon Pocket Park Priors Hall West Regionally Important Geological and Geomorpological Site Weldon Woodland Site Regionally Important Geological and Geomorphological Site Species lists and descriptions for most of these non-statutory sites are attached to this report. Information concerning the RIGG sites has not been digitised but hardcopies of citations are available if necessary. Species records 39 protected and notable species records fall within your specified search boundaries. A list of these species records is attached to this report. -

Phytosociological Analysis of Natural and Artificial Pine Forests Of
Article Phytosociological Analysis of Natural and Artificial Pine Forests of the Class Vaccinio-Piceetea Br.-Bl. in Br.-Bl. et al. 1939 in the Sudetes and Their Foreland (Bohemian Massif, Central Europe) Kamila Reczy ´nska 1 , Paweł Pech 2 and Krzysztof Swierkosz´ 3,* 1 Department of Botany, Institute of Environmental Biology, University of Wrocław, Kanonia 6/8, PL-50-328 Wrocław, Poland; [email protected] 2 Bureau of Forest Management and Geodesy, Piastowska 9, PL-49-300 Brzeg, Poland; [email protected] 3 Museum of Natural History, Faculty of Biological Sciences, University of Wrocław, Sienkiewicza 21, PL-50-335 Wrocław, Poland * Correspondence: [email protected] Abstract: Research Highlights: Differentiation of Scots pine forests of the class Vaccinio-Piceetea in Poland has been the subject of numerous studies, including revisions. Despite that, the area of southwestern Poland was hitherto practically unexplored in this respect. Background and Objectives: The aim of this work was therefore (i) to present the diversity of the pine forests in the Sudetes and their foreland; (ii) to compare the ecology of studied communities. Materials and Methods: We analyzed 175 phytosociological relevés collected between 1991 and 2020 in natural and anthropogenic pine stands. To identify vegetation types, we used the modified TWINSPAN algorithm; principal coordinate analysis, distance-based redundancy analysis and permutational tests were applied to identify the variation explained and the main environmental gradients shaping the studied plant com- munities. Results: Five associations were distinguished: thermophilous Asplenio cuneifolii-Pinetum Citation: Reczy´nska,K.; Pech, P.; sylvestris Pišta ex Husová in Husová et al. -

Slender False Brome (Brachypodium Sylvaticum, Poaceae), an Invasive
Slender False Brome ( Brachypodium sylvaticum , Poaceae), an Invasive Grass New to Ontario, Canada BRIAN M. M ILLER 1, ROBERT J. A ITKEN 1, MICHAEL J. O LDHAM 2, and ANTON A. R EZNICEK 3 1 Aboud & Associates Inc. ( Consulting Arborists, Ecologists and Landscape Designers) , 591 Woolwich Street, Guelph, Ontario N1H 3Y5 Canada; email: [email protected] and [email protected] 2 Ontario Natural Heritage Information Centre, Ontario Ministry of Natural Resources, 300 Water Street, Peterborough, Ontario K9L 1C8 Canada; email: [email protected] 3 University of Michigan Herbarium, 3600 Varsity Drive, Ann Arbor, Michigan 48108-2287 USA; email: [email protected] Miller, Brian M., Robert J. Aitken, Michael J. Oldham, and Anton A. Reznicek. 2011. Slender False Brome ( Brachypodium sylvaticum , Poaceae), an invasive grass new to Ontario, Canada. Canadian Field-Naturalist 125(3): 235 –240. Brachypodium sylvaticum , Slender False Brome, an invasive Eurasian grass, is reported for the first time in Ontario and eastern Canada from Grey County, southern Ontario. The only previous Canadian record is from Vancouver Island, British Columbia. The species is widespread in the U.S. Pacific Northwest, where it is spreading aggressively throughout much of western Oregon. In the eastern U.S.A., known populations are few and localized, although the species will likely spread. Key Words: Brachypodium sylvaticum , Slender False Brome, Poaceae, invasive, new record, Grey County, Ontario, Canada. The invasive Eurasian grass species Brachypodium America, it was subsequently discovered in San Mateo sylvaticum (Huds.) P. Beauv. (Slender False Brome) County, California, in 2003 (Johnson 2004) and near was recently discovered in Ontario, and is here report - Cowichan Lake, Vancouver Island, British Columbia, ed as new to the flora of the province and eastern Cana - in 2008 (Fenneman 2010*). -
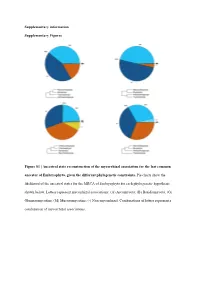
Ancestral State Reconstruction of the Mycorrhizal Association for the Last Common Ancestor of Embryophyta, Given the Different Phylogenetic Constraints
Supplementary information Supplementary Figures Figure S1 | Ancestral state reconstruction of the mycorrhizal association for the last common ancestor of Embryophyta, given the different phylogenetic constraints. Pie charts show the likelihood of the ancestral states for the MRCA of Embryophyta for each phylogenetic hypothesis shown below. Letters represent mycorrhizal associations: (A) Ascomycota; (B) Basidiomycota; (G) Glomeromycotina; (M) Mucoromycotina; (-) Non-mycorrhizal. Combinations of letters represent a combination of mycorrhizal associations. Austrocedrus chilensis Chamaecyparis obtusa Sequoiadendron giganteum Prumnopitys taxifolia Prumnopitys Prumnopitys montana Prumnopitys Prumnopitys ferruginea Prumnopitys Araucaria angustifolia Araucaria Dacrycarpus dacrydioides Dacrycarpus Taxus baccata Podocarpus oleifolius Podocarpus Afrocarpus falcatus Afrocarpus Ephedra fragilis Nymphaea alba Nymphaea Gnetum gnemon Abies alba Abies balsamea Austrobaileya scandens Austrobaileya Abies nordmanniana Thalictrum minus Thalictrum Abies homolepis Caltha palustris Caltha Abies magnifica ia repens Ranunculus Abies religiosa Ranunculus montanus Ranunculus Clematis vitalba Clematis Keteleeria davidiana Anemone patens Anemone Tsuga canadensis Vitis vinifera Vitis Tsuga mertensiana Saxifraga oppositifolia Saxifraga Larix decidua Hypericum maculatum Hypericum Larix gmelinii Phyllanthus calycinus Phyllanthus Larix kaempferi Hieronyma oblonga Hieronyma Pseudotsuga menziesii Salix reinii Salix Picea abies Salix polaris Salix Picea crassifolia Salix herbacea -

A Guide to North American Grasslands
Desert Volume 29, Number 2 Published by The University of Arizona for Plants the Boyce Thompson Arboretum A Guide to North American Grasslands David E. Brown and Elizabeth Makings Relict Great Basin Shrub-Grassland near Wupatki National Monument northeast of Flagstaff, Coconino County, Arizona, 1,650 m (5,413 ft). Volume 29, Number 2 Desert Plants Published by The University of Arizona for the Boyce Thompson Arboretum A journal devoted to broadening knowledge of plants 37615 E US Highway 60 indigenous or adapted to arid and sub-arid regions and Superior, AZ 85173 to encouraging the appreciation of these plants. Copyright 2014. The Arizona Board of Regents on Mark D. Siegwarth, editor behalf of The University of Arizona. The Boyce [email protected] Thompson Arboretum at Superior, Arizona, is cooperatively managed by the Boyce Thompson Production Director: Kim Stone Southwestern Arboretum, Inc., The University of Arizona, and Arizona State Parks. Boyce Thompson Arboretum From the editor As Desert Plants begins its 35th year with a new staff, it in editorship comes the opportunity to rethink what Desert seems somewhat appropriate that as we begin a new chap- Plants is and could be. Desert Plants is devoted to broad- ter in the history of Desert Plants, we start with A Guide ening knowledge of plants indigenous or adapted to arid to North American Grasslands by David E. Brown and Eliza- and sub-arid regions and to encouraging the appreciation of beth Makings. Probably one of the most quoted, used and these plants. With such a broad mandate, it is open to vari- reprinted issues of Desert Plants to this day is Volume 4, ous interpretations. -

Sea Wall Biodiversity Handbook by Tim Gardiner, Rob Pilcher and Max Wade
Sea Wall Biodiversity Handbook Sea Wall Tim Gardiner, Biodiversity Officer at the Environment Agency, Rob Pilcher, Ecology Team Leader for North West England at AECOM and Max Wade, Technical Director (Ecology) at AECOM, have a long standing interest in the ecology and management of sea wall habitats. Their handbook on sea wall biodiversity brings together a wealth of knowledge about this Cinderella habitat based on the authors’ experience of practical management and the flora and fauna of sea walls. The handbook highlights the breadth of plant and animal species living and relying on sea walls and provides practical guidance for managers of sea defences to ensure that their biodiversity value is by conserved and enhanced. Tim Gardiner, Rob Pilcher and Max Wade Rob Pilcher Gardiner, Tim Sea Wall Biodiversity Handbook by Tim Gardiner, Rob Pilcher and Max Wade SeaWall Layout Cvr v1.indd 1 02/09/2015 15:09 SeaWall Layout Txt.indd 4 20/08/2015 15:57 Sea Wall Biodiversity Handbook Sea Wall Biodiversity Handbook by Tim Gardiner, Rob Pilcher & Max Wade © Copyright First published in 2015 by RPS Images are the authors unless labelled. Designed and Printed by Mimeo Limited Units 1-3, The Ermine Centre, Hurricane Close, Huntingdon, Cambridgeshire PE29 6XX. A CIP record is available from the British Lending Library in London. ISBN: 978-0-9546600-4-8 Citation; Gardiner, T., Pilcher, R. & Wade, M. (2015) Sea Wall Biodiversity Handbook. RPS. SeaWall Layout Txt.indd 3 20/08/2015 15:57 SeaWall Layout Txt.indd 4 20/08/2015 15:57 Sea Wall Biodiversity Handbook Acknowledgements Thanks go to those involved with preparation of the case studies, to site managers and their respective organisations for allowing data to be used and for reviewing draft text of the case studies.