Reconstructed Skin Models Revealed Unexpected Differences In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Critical Evaluation of Gene Expression Changes in Human Tissues In
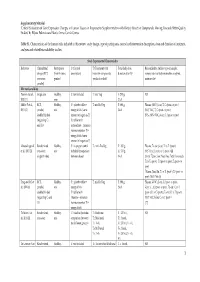
Supplementary Material ‘Critical Evaluation of Gene Expression Changes in Human Tissues in Response to Supplementation with Dietary Bioactive Compounds: Moving Towards Better-Quality Studies’ by Biljana Pokimica and María-Teresa García-Conesa Table S1. Characteristics of the human trials included in this review: study design, type of participants, control and intervention description, dose and duration of treatment, analyses and related bioavailability studies. Study Experimental Characteristics Reference Clinical trial Participants C (Control T (Treatment with Total daily dose, Bioavailability studies: type of sample, design (RCT, (health status, description) bioactive compounds, duration (d or h)1 compounds and (or) metabolites analysed, crossover, gender) products or diet) main results2 parallel) Mix meals and diets Persson I et al., Single arm Healthy, C: not included T: mix Veg T: 250 g, NR 2000 [1] men 21 d Møller P et al., RCT, Healthy, C1: placebo tablet + T: mix FruVeg T: 600 g, Plasma: (NS↑) β-car, T, C2 (post- vs pre-) 2003 [2] parallel, mix energy drink (same 24 d (NC) VitC, T, C2 (post- vs pre-) double blinded amount of sugars as T) (NS↓, 69%) VitC, β-car, C1 (post- vs pre-) (regarding C1 C2: tablet with and C2) antioxidants + minerals (same amount as T) + energy drink (same amount of sugars as T) Almendingen K Randomized, Healthy, C: no proper control T1,2: mix FruVeg T1: 300 g, Plasma: ↑α-car, β-car, T2 vs T1 (post-) et al., 2005 [3] crossover, mix included (comparison T2: 750 g, (NS↑) Lyc, Lut, T2 vs T1 (post-) [4] single -

Microarray Analysis of Novel Genes Involved in HSV- 2 Infection
Microarray analysis of novel genes involved in HSV- 2 infection Hao Zhang Nanjing University of Chinese Medicine Tao Liu ( [email protected] ) Nanjing University of Chinese Medicine https://orcid.org/0000-0002-7654-2995 Research Article Keywords: HSV-2 infection,Microarray analysis,Histospecic gene expression Posted Date: May 12th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-517057/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/19 Abstract Background: Herpes simplex virus type 2 infects the body and becomes an incurable and recurring disease. The pathogenesis of HSV-2 infection is not completely clear. Methods: We analyze the GSE18527 dataset in the GEO database in this paper to obtain distinctively displayed genes(DDGs)in the total sequential RNA of the biopsies of normal and lesioned skin groups, healed skin and lesioned skin groups of genital herpes patients, respectively.The related data of 3 cases of normal skin group, 4 cases of lesioned group and 6 cases of healed group were analyzed.The histospecic gene analysis , functional enrichment and protein interaction network analysis of the differential genes were also performed, and the critical components were selected. Results: 40 up-regulated genes and 43 down-regulated genes were isolated by differential performance assay. Histospecic gene analysis of DDGs suggested that the most abundant system for gene expression was the skin, immune system and the nervous system.Through the construction of core gene combinations, protein interaction network analysis and selection of histospecic distribution genes, 17 associated genes were selected CXCL10,MX1,ISG15,IFIT1,IFIT3,IFIT2,OASL,ISG20,RSAD2,GBP1,IFI44L,DDX58,USP18,CXCL11,GBP5,GBP4 and CXCL9.The above genes are mainly located in the skin, immune system, nervous system and reproductive system. -

Differentially Expressed Late Constituents of the Epidermal Cornified Envelope
Differentially expressed late constituents of the epidermal cornified envelope D. Marshall, M. J. Hardman, K. M. Nield, and C. Byrne* School of Biological Sciences, University of Manchester, 3.239 Stopford Building, Oxford Road, Manchester M13 9PT, United Kingdom Communicated by Elaine Fuchs, University of Chicago, Chicago, IL, September 17, 2001 (received for review June 23, 2001) Barrier activity of skin and internal barrier-forming epithelial There have been persistent reports of additional genes͞ linings are conferred by a lipid-corneocyte structure (stratum proteins with structures homologous to cornified envelope pro- corneum in skin).The integrity of the corneocytes depends on the teins. These include XP5, XP31, XP32 (12), the newly identified outer cornified envelope and is essential for maintenance of barrier component of the EDC (NICE-1; ref. 13), protein products of a function. During epidermal development and differentiation, pro- range of annotated expressed sequence tags (ESTs; ref. 14), and teins are sequentially incorporated into the envelope via action of Eig3 protein (15). We show that at least some of these proteins epidermal transglutaminases in a well documented process. How- are encoded by a previously undetected gene cluster in the ever, recent knockouts of major cornified envelope constituents human epidermal differentiation complex (EDC) at 1q21 (16), have failed to disrupt barrier function significantly, suggesting that with homologues detected in mouse (12, 14, 16). We show that additional unidentified components are involved. We report a new these genes encode proteins, which are new cornified envelope gene cluster in the epidermal differentiation complex at human constituents distinct from SPRRs. Like SPRRs, the genes are 1q21 encoding a family of 18 proteins that are substrates for differentially expressed in different types of barrier epithelia. -

University of California, San Diego
UC San Diego UC San Diego Electronic Theses and Dissertations Title The post-terminal differentiation fate of RNAs revealed by next-generation sequencing Permalink https://escholarship.org/uc/item/7324r1rj Author Lefkowitz, Gloria Kuo Publication Date 2012 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA, SAN DIEGO The post-terminal differentiation fate of RNAs revealed by next-generation sequencing A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Biomedical Sciences by Gloria Kuo Lefkowitz Committee in Charge: Professor Benjamin D. Yu, Chair Professor Richard Gallo Professor Bruce A. Hamilton Professor Miles F. Wilkinson Professor Eugene Yeo 2012 Copyright Gloria Kuo Lefkowitz, 2012 All rights reserved. The Dissertation of Gloria Kuo Lefkowitz is approved, and it is acceptable in quality and form for publication on microfilm and electronically: __________________________________________________________________ __________________________________________________________________ __________________________________________________________________ __________________________________________________________________ __________________________________________________________________ Chair University of California, San Diego 2012 iii DEDICATION Ma and Ba, for your early indulgence and support. Matt and James, for choosing more practical callings. Roy, my love, for patiently sharing the ups and downs -

Evaluation on Reprogramed Biological Processes in Transgenic Maize
www.nature.com/scientificreports OPEN Evaluation on reprogramed biological processes in transgenic maize varieties using transcriptomics and metabolomics Wei Fu1, Pengyu Zhu1, Mingnan Qu2, Wang Zhi1, Yongjiang Zhang1, Feiwu Li3* & Shuifang Zhu1* Genetic engineering (GM) has great potential to improve maize productivity, but rises some concerns on unintended efects, and equivalent as their comparators. There are some limitations through targeted analysis to detect the UE in genetically modifed organisms in many previous studies. We here reported a case-study on the efects of introducing herbicides and insect resistance (HIR) gene cassette on molecular profling (transcripts and metabolites) in a popular maize variety Zhengdan958 (ZD958) in China. We found that introducing HIR gene cassette bring a limited numbers of diferential abundant genes (DAGs) or diferential abundant metabolites (DAMs) between transgenic events and non-transgenic control. In contrast, averaged 10 times more DAGs and DAMs were observed when performed comparison under diferent growing environments in three diferent ecological regions of China than the numbers induced by gene efects. Major biological pathways relating to stress response or signaling transduction could explain somehow the efects of growing environments. We further compared two transgenic events mediated ZD958 (GM-ZD958) with either transgenic parent GM-Z58, and other genetic background nonGM-Z58, nonGM-ZD958, and Chang7-2. We found that the numbers of DAGs and DAMs between GM-ZD958 and its one parent maize variety, Z58 or GM-Z58 is equivalent, but not Chang7-2. These fndings suggest that greater efects due to diferent genetic background on altered molecular profling than gene modifcation itself. This study provides a case evidence indicating marginal efects of gene pleiotropic efects, and environmental efects should be emphasized. -

Cystatin-B Negatively Regulates the Malignant Characteristics of Oral Squamous Cell Carcinoma Possibly Via the Epithelium Proliferation/ Differentiation Program
ORIGINAL RESEARCH published: 24 August 2021 doi: 10.3389/fonc.2021.707066 Cystatin-B Negatively Regulates the Malignant Characteristics of Oral Squamous Cell Carcinoma Possibly Via the Epithelium Proliferation/ Differentiation Program Tian-Tian Xu 1, Xiao-Wen Zeng 1, Xin-Hong Wang 2, Lu-Xi Yang 1, Gang Luo 1* and Ting Yu 1* 1 Department of Periodontics, Affiliated Stomatology Hospital of Guangzhou Medical University, Guangzhou Key Laboratory of Basic and Applied Research of Oral Regenerative Medicine, Guangzhou, China, 2 Department of Oral Pathology and Medicine, Affiliated Stomatology Hospital of Guangzhou Medical University, Guangzhou Key Laboratory of Basic and Applied Research of Oral Regenerative Medicine, Guangzhou, China Disturbance in the proteolytic process is one of the malignant signs of tumors. Proteolysis Edited by: Eva Csosz, is highly orchestrated by cysteine cathepsin and its inhibitors. Cystatin-B (CSTB) is a University of Debrecen, Hungary general cysteine cathepsin inhibitor that prevents cysteine cathepsin from leaking from Reviewed by: lysosomes and causing inappropriate proteolysis. Our study found that CSTB was Csongor Kiss, downregulated in both oral squamous cell carcinoma (OSCC) tissues and cells University of Debrecen, Hungary Gergely Nagy, compared with normal controls. Immunohistochemical analysis showed that CSTB was University of Debrecen, Hungary mainly distributed in the epithelial structure of OSCC tissues, and its expression intensity *Correspondence: was related to the grade classification. A correlation analysis between CSTB and clinical Gang Luo [email protected] prognosis was performed using gene expression data and clinical information acquired Ting Yu from The Cancer Genome Atlas (TCGA) database. Patients with lower expression levels of [email protected] CSTB had shorter disease-free survival times and poorer clinicopathological features Specialty section: (e.g., lymph node metastases, perineural invasion, low degree of differentiation, and This article was submitted to advanced tumor stage). -

Analysis of the Ultraviolet B Response in Primary Human Keratinocytes Using Oligonucleotide Microarrays
Analysis of the ultraviolet B response in primary human keratinocytes using oligonucleotide microarrays Angela Sesto*, Manuel Navarro*†, Frank Burslem‡, and Jose´L. Jorcano* *Department of Molecular and Cell Biology, Centro de Investigaciones Energe´ticas, Medioambientales y Tecnolo´gicas (CIEMAT), Avenida Complutense 22, 28040 Madrid, Spain; and ‡Pfizer Global Research and Development, Sandwich, Kent CT13 9NJ, United Kingdom Communicated by A. Garcia-Bellido, Autonomous University of Madrid, Madrid, Spain, December 17, 2001 (received for review October 19, 2001) UV radiation is the most important environmental skin aggressor, To understand the mechanisms of UV response, we sought to causing cancer and other problems. This paper reports the use of obtain the transcriptional profile of primary keratinocytes after oligonucleotide microarray technology to determine changes in UVB irradiation. Cultured human epidermal keratinocytes were gene expression in human keratinocytes after UVB treatment. treated with three UV doses, and samples were collected at Examination of the effects of different doses at different times different intervals. Oligonucleotide microarrays containing over after irradiation gave a global picture of the keratinocyte response 6,000 genes (HuGeneFL, Affymetrix, Santa Clara, CA) were to this type of insult. Five hundred thirty-nine regulated transcripts used to quantitatively assess changes in gene expression. The were found and organized into nine different clusters depending subset of regulated transcripts was classified into self-organizing on behavior patterns. Classification of these genes into 23 func- maps according to their different behaviors. Biological functions tional categories revealed that several biological processes are were assigned to these genes, which were then correlated with globally affected by UVB. -

393LN V 393P 344SQ V 393P Probe Set Entrez Gene
393LN v 393P 344SQ v 393P Entrez fold fold probe set Gene Gene Symbol Gene cluster Gene Title p-value change p-value change chemokine (C-C motif) ligand 21b /// chemokine (C-C motif) ligand 21a /// chemokine (C-C motif) ligand 21c 1419426_s_at 18829 /// Ccl21b /// Ccl2 1 - up 393 LN only (leucine) 0.0047 9.199837 0.45212 6.847887 nuclear factor of activated T-cells, cytoplasmic, calcineurin- 1447085_s_at 18018 Nfatc1 1 - up 393 LN only dependent 1 0.009048 12.065 0.13718 4.81 RIKEN cDNA 1453647_at 78668 9530059J11Rik1 - up 393 LN only 9530059J11 gene 0.002208 5.482897 0.27642 3.45171 transient receptor potential cation channel, subfamily 1457164_at 277328 Trpa1 1 - up 393 LN only A, member 1 0.000111 9.180344 0.01771 3.048114 regulating synaptic membrane 1422809_at 116838 Rims2 1 - up 393 LN only exocytosis 2 0.001891 8.560424 0.13159 2.980501 glial cell line derived neurotrophic factor family receptor alpha 1433716_x_at 14586 Gfra2 1 - up 393 LN only 2 0.006868 30.88736 0.01066 2.811211 1446936_at --- --- 1 - up 393 LN only --- 0.007695 6.373955 0.11733 2.480287 zinc finger protein 1438742_at 320683 Zfp629 1 - up 393 LN only 629 0.002644 5.231855 0.38124 2.377016 phospholipase A2, 1426019_at 18786 Plaa 1 - up 393 LN only activating protein 0.008657 6.2364 0.12336 2.262117 1445314_at 14009 Etv1 1 - up 393 LN only ets variant gene 1 0.007224 3.643646 0.36434 2.01989 ciliary rootlet coiled- 1427338_at 230872 Crocc 1 - up 393 LN only coil, rootletin 0.002482 7.783242 0.49977 1.794171 expressed sequence 1436585_at 99463 BB182297 1 - up 393 -

Non‑Invasive Proteome‑Wide Quantification of Skin Barrier‑Related Proteins Using Label‑Free LC‑MS/MS Analysis
MOLECULAR MEDICINE REPORTS 21: 2227-2235, 2020 Non‑invasive proteome‑wide quantification of skin barrier‑related proteins using label‑free LC‑MS/MS analysis MENGTING LIU1,2*, JING ZHANG1,2*, YAOCHI WANG1,2, CONG XIN1,2, JIE MA1,2, SHUANGJUN XU1,2, XIAOMENG WANG1,2, JINPING GAO1,2, XUEJUN ZHANG1,2 and SEN YANG1,2 1Department of Dermatology, The First Affiliated Hospital, Anhui Medical University;2 Key Laboratory of Dermatology, Anhui Medical University, Ministry of Education, Hefei, Anhui 230032, P.R. China Received August 6, 2019; Accepted February 6, 2020 DOI: 10.3892/mmr.2020.11020 Abstract. A number of epidermal proteins are closely related invasion (1,2). If the barrier integrity or function is disrupted, to skin barrier function, the abnormalities of which can lead water in the dermis and epidermis will be lost, the balance to specific skin diseases. These proteins must be quantified of the skin microecology will be disrupted (3), and hostile to further investigate the changes in the skin barrier between factors from the external environment will easily invade the healthy and disease states. However, the non‑invasive and skin, leading to skin ageing and the occurrence of various skin proteome‑wide quantification of skin proteins without any diseases (1,2,4). labelling steps remains a challenge. In this study, 3M medical Numerous functions of the skin barrier, including struc- adhesive tapes were used to obtain skin samples from tural, regulatory, hygroscopic and signalling functions, rely volunteers. Proteins were extracted from fresh skin samples on epidermal proteins (5,6). Keratins, cytoskeletal proteins and digested with trypsin. -

A Comprehensive Data Analysis of Differentially Regulated Genes in Melanoma
A Comprehensive Data Analysis of Differentially Regulated Genes in Melanoma Yanjie Han ( [email protected] ) Clinical Laboratory, Kaifeng Central Hospital, 85 Hedao Street, Kaifeng, Henan 475000, China. https://orcid.org/0000-0003-3755-1386 Xinxin Li Functional Laboratory, Kaifeng Central Hospital, Kaifeng, Henan 475000 Jiliang Yan Clinical Laboratory,Kaifeng Central Hospital, Kaifeng, Henan 475000 Chunyan Ma Clinical Laboratory,Kaifeng Central Hospital, Kaifeng, Henan 475000 Xin Wang Department of Stomatology, Kaifeng Central Hospital, Kaifeng, Henan 475000 Hong Pan Department of Stomatology, Kaifeng Central Hospital, Kaifeng, Henan 475000 Xiaoli Zheng Hospital Infection Control Oce, First Aliated Hospital of Henan University, Kaifeng, Henan 475000 Zhen Zhang Clinical Laboratory,Kaifeng Central Hospital, Kaifeng, Henan 475000 Biao Gao Clinical Laboratory,Kaifeng Central Hospital, Kaifeng, Henan 475000 Xin-Ying JI ( [email protected] ) Kaifeng Key Laboratory for Infectious Diseases and Biosafety, Henan International Joint Laboratory of Nuclear Protein Regulation, Henan University College of Medicine, Kaifeng, Henan 475004 Research Keywords: melanoma, DEGs, bioinformatics analysis Posted Date: March 30th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-19035/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/16 Abstract Background: Melanoma is the most deadly tumor in skin tumors and is prone to distant metastases. The incidence of melanoma has increased rapidly in the past few decades, and current trends indicate that this growth is continuing. This study was aimed to explore the molecular mechanisms of melanoma pathogenesis and discover underlying pathways and genes associated with melanoma. Methods: We used high-throughput expression data to study differential expression proles of related genes in melanoma. -

Genes Encoding Structural Proteins of Epidermal Cornification
Genes Encoding Structural Proteins of Epidermal Cornification and S100 Calcium-Binding Proteins Form a Gene Complex ("Epidermal Differentiation Complex") on Human Chromosome lq21 Dietmar Mischke,* Bernhard P. Korge,t Ingo Marenholz,* Armin Volz,* and Andreas Ziegler* *Insti tut flir Experimente llc Onkologie und Transplantationsmedizin, Virchow-Kliniku111 der Hutllbold t-Univc rsitiit ZlI Berl in. Berlin; and t Klinik lind Poliklinik fUr Demlatologic und Venerologie, Universitii t Z lI Knln. Knln. Gemlan), Chromosome 1 reveals in region lq21 a most remark chohyalin, profilaggrin, involucrin, SPRR3, SPRRIB, able density of genes that fulfill important functions SPRR2A, loricrin, S100A9, StOOA8, StOOA6. Co in terminal differentiation of the human epidermis. localization of genes expressed late during matura These genes encode the cornified envelope precur tion of epidermal cells together with genes encodin g sors loricrin, involucrin, and small proline-rich pro calcium-binding proteins is particularly intriguing teins (SPRRt, SPRR2, and SPRR3), the intermediate since calciulll levels tightly control the differe ntia filament-associated proteins profilaggrin and tri tion of epithelial cells and the expression of genes chohyalin, and several S100A calciUln-binding pro encoding epidermal structural proteins. Accounting teins. Extending and refining our previous physical for the close functional cooperation alllong the se tnap of lq21 we have now mapped two additional structurally and evolutionary related genes, we con S100A genes as well as the three SPRR subfamilies clude that these loci constitute a gene cOlllplex, for and resolved the arrangement of involucrin, SPRRs, which we propose the name epidermal differentiation and loricrin. All genes are linked within 1.9 Mbp of complex. -

AKT1-Mediated Lamin A&Sol;C Degradation Is Required for Nuclear
Cell Death and Differentiation (2015) 22, 2123–2132 & 2015 Macmillan Publishers Limited All rights reserved 1350-9047/15 www.nature.com/cdd AKT1-mediated Lamin A/C degradation is required for nuclear degradation and normal epidermal terminal differentiation AS Naeem1,2, Y Zhu1,2,WLDi1,2, S Marmiroli3 and RFL O’Shaughnessy*,1,2 Nuclear degradation is a key stage in keratinocyte terminal differentiation and the formation of the cornified envelope that comprises the majority of epidermal barrier function. Parakeratosis, the retention of nuclear material in the cornified layer of the epidermis, is a common histological observation in many skin diseases, notably in atopic dermatitis and psoriasis. Keratinocyte nuclear degradation is not well characterised, and it is unclear whether the retained nuclei contribute to the altered epidermal differentiation seen in eczema and psoriasis. Loss of AKT1 function strongly correlated with parakeratosis both in eczema samples and in organotypic culture models. Although levels of DNAses, including DNase1L2, were unchanged, proteomic analysis revealed an increase in Lamin A/C. AKT phosphorylates Lamin A/C, targeting it for degradation. Consistent with this, Lamin A/C degradation was inhibited and Lamin A/C was observed in the cornified layer of AKT1 knockdown organotypic cultures, surrounding retained nuclear material. Using AKT-phosphorylation-dead Lamin A constructs we show that the retention of nuclear material is sufficient to cause profound changes in epidermal terminal differentiation, specifically a reduction in Loricrin, Keratin 1, Keratin 10, and filaggrin expression. We show that preventing nuclear degradation upregulates BMP2 expression and SMAD1 signalling. Consistent with these data, we observe both parakeratosis and evidence of increased SMAD1 signalling in atopic dermatitis.