PML Regulates the Epidermal Differentiation Complex and Skin
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Critical Evaluation of Gene Expression Changes in Human Tissues In
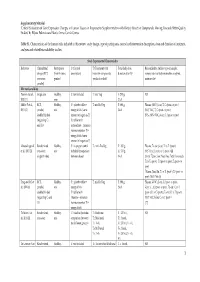
Supplementary Material ‘Critical Evaluation of Gene Expression Changes in Human Tissues in Response to Supplementation with Dietary Bioactive Compounds: Moving Towards Better-Quality Studies’ by Biljana Pokimica and María-Teresa García-Conesa Table S1. Characteristics of the human trials included in this review: study design, type of participants, control and intervention description, dose and duration of treatment, analyses and related bioavailability studies. Study Experimental Characteristics Reference Clinical trial Participants C (Control T (Treatment with Total daily dose, Bioavailability studies: type of sample, design (RCT, (health status, description) bioactive compounds, duration (d or h)1 compounds and (or) metabolites analysed, crossover, gender) products or diet) main results2 parallel) Mix meals and diets Persson I et al., Single arm Healthy, C: not included T: mix Veg T: 250 g, NR 2000 [1] men 21 d Møller P et al., RCT, Healthy, C1: placebo tablet + T: mix FruVeg T: 600 g, Plasma: (NS↑) β-car, T, C2 (post- vs pre-) 2003 [2] parallel, mix energy drink (same 24 d (NC) VitC, T, C2 (post- vs pre-) double blinded amount of sugars as T) (NS↓, 69%) VitC, β-car, C1 (post- vs pre-) (regarding C1 C2: tablet with and C2) antioxidants + minerals (same amount as T) + energy drink (same amount of sugars as T) Almendingen K Randomized, Healthy, C: no proper control T1,2: mix FruVeg T1: 300 g, Plasma: ↑α-car, β-car, T2 vs T1 (post-) et al., 2005 [3] crossover, mix included (comparison T2: 750 g, (NS↑) Lyc, Lut, T2 vs T1 (post-) [4] single -

Differential Roles of P63 Isoforms in Epidermal Development: Selective Genetic Complementation in P63 Null Mice
Cell Death and Differentiation (2006) 13, 1037–1047 & 2006 Nature Publishing Group All rights reserved 1350-9047/06 $30.00 www.nature.com/cdd Differential roles of p63 isoforms in epidermal development: selective genetic complementation in p63 null mice E Candi1, A Rufini1, A Terrinoni1, D Dinsdale2, M Ranalli1, The skin consists of two compartments, the dermis and the A Paradisi1, V De Laurenzi2, LG Spagnoli1, MV Catani1, epidermis. The latter is a multilayered, stratified epithelium S Ramadan1, RA Knight2 and G Melino*,1,2 continuously regenerated by terminally differentiating keratino- cytes, a process called cornification1–3 (Figure 1a and b). 4 1 Biochemistry Laboratory, IDI-IRCCS, c/o University of Rome ‘Tor Vergata’, Recent evidence demonstrates a major role for p63, a 00133 Rome, Italy member of the p53 family,5–8 in this process as mutations in 2 Medical Research Council, Toxicology Unit, Leicester University, Leicester the TP63 gene cause limb and skin defects in humans,9 and LE1 9HN, UK p63À/À mice have no epidermis, no limbs and die at birth * Corresponding author: G Melino, Medical Research Council, Toxicology Unit, owing to dehydration.10,11 Hodgkin Building, Leicester University, Lancaster Road, PO Box 138, Leicester LE1 9HN, UK. Tel: þ 44 116 252 5616; Fax: þ 44 116 252 5551; The expression of p63 proteins originates from two E-mail: [email protected] promoters, giving rise to TAp63 and DNp63 isoforms. In addition, both isoforms undergo alternative splicing at the Received 03.3.06; revised 08.3.06; accepted 08.3.06; published online 07.4.06 C-terminus producing three different TAp63 and DNp63 Edited by P Vandenabeele isoforms (a, b and g). -

Cell Shape Controls Terminal Differentiation of Human Epidermal Keratinocytes (Cell Adhesion/Proliferation/Involucrin Expression) FIONA M
Proc. Natl. Acad. Sci. USA Vol. 85, pp. 5576-5580, August 1988 Cell Biology Cell shape controls terminal differentiation of human epidermal keratinocytes (cell adhesion/proliferation/involucrin expression) FIONA M. WATT*, PETER W. JORDANt, AND CHARLES H. O'NEILLt *Keratinocyte Laboratory, and tAnchorage Laboratory, Imperial Cancer Research Fund, P.O. Box 123, Lincoln's Inn Fields, London WC2A 3PX, United Kingdom Communicated by Howard Green, May 2, 1988 ABSTRACT Cultures of human epidermal keratinocytes To investigate the role of cell-substratum contact, and provide a useful experimental model with which to study the hence cell shape, in regulating terminal differentiation, we factors that regulate cell proliferation and terminal differen- have made use of a recently described technique that allows tiation. One situation that is known to trigger premature precise control of cell-substratum contact in the absence of terminal differentiation is suspension culture, when keratin- cell-cell contact (14). We have looked at the effect ofchanges ocytes are deprived of substratum and intercellular contact. in cell shape on DNA synthesis and involucrin expression We have now investigated whether area ofsubstratum contact, and conclude that a rounded morphology acts as a signal to and hence cell shape, can regulate terminal differentiation. keratinocytes to stop dividing and to undergo terminal Keratinocytes were grown on circular adhesive islands that differentiation; inhibition ofproliferation in spread cells is not prevented cell-cell contact. By varying island area we could a sufficient signal for terminal differentiation. vary cell shape from fully spread to almost spherical. We found that when substratum contact was restricted, DNA synthesis was inhibited and expression of involucrin, a marker of MATERIALS AND METHODS terminal differentiation, was stimulated. -

Microarray Analysis of Novel Genes Involved in HSV- 2 Infection
Microarray analysis of novel genes involved in HSV- 2 infection Hao Zhang Nanjing University of Chinese Medicine Tao Liu ( [email protected] ) Nanjing University of Chinese Medicine https://orcid.org/0000-0002-7654-2995 Research Article Keywords: HSV-2 infection,Microarray analysis,Histospecic gene expression Posted Date: May 12th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-517057/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/19 Abstract Background: Herpes simplex virus type 2 infects the body and becomes an incurable and recurring disease. The pathogenesis of HSV-2 infection is not completely clear. Methods: We analyze the GSE18527 dataset in the GEO database in this paper to obtain distinctively displayed genes(DDGs)in the total sequential RNA of the biopsies of normal and lesioned skin groups, healed skin and lesioned skin groups of genital herpes patients, respectively.The related data of 3 cases of normal skin group, 4 cases of lesioned group and 6 cases of healed group were analyzed.The histospecic gene analysis , functional enrichment and protein interaction network analysis of the differential genes were also performed, and the critical components were selected. Results: 40 up-regulated genes and 43 down-regulated genes were isolated by differential performance assay. Histospecic gene analysis of DDGs suggested that the most abundant system for gene expression was the skin, immune system and the nervous system.Through the construction of core gene combinations, protein interaction network analysis and selection of histospecic distribution genes, 17 associated genes were selected CXCL10,MX1,ISG15,IFIT1,IFIT3,IFIT2,OASL,ISG20,RSAD2,GBP1,IFI44L,DDX58,USP18,CXCL11,GBP5,GBP4 and CXCL9.The above genes are mainly located in the skin, immune system, nervous system and reproductive system. -

Differentially Expressed Late Constituents of the Epidermal Cornified Envelope
Differentially expressed late constituents of the epidermal cornified envelope D. Marshall, M. J. Hardman, K. M. Nield, and C. Byrne* School of Biological Sciences, University of Manchester, 3.239 Stopford Building, Oxford Road, Manchester M13 9PT, United Kingdom Communicated by Elaine Fuchs, University of Chicago, Chicago, IL, September 17, 2001 (received for review June 23, 2001) Barrier activity of skin and internal barrier-forming epithelial There have been persistent reports of additional genes͞ linings are conferred by a lipid-corneocyte structure (stratum proteins with structures homologous to cornified envelope pro- corneum in skin).The integrity of the corneocytes depends on the teins. These include XP5, XP31, XP32 (12), the newly identified outer cornified envelope and is essential for maintenance of barrier component of the EDC (NICE-1; ref. 13), protein products of a function. During epidermal development and differentiation, pro- range of annotated expressed sequence tags (ESTs; ref. 14), and teins are sequentially incorporated into the envelope via action of Eig3 protein (15). We show that at least some of these proteins epidermal transglutaminases in a well documented process. How- are encoded by a previously undetected gene cluster in the ever, recent knockouts of major cornified envelope constituents human epidermal differentiation complex (EDC) at 1q21 (16), have failed to disrupt barrier function significantly, suggesting that with homologues detected in mouse (12, 14, 16). We show that additional unidentified components are involved. We report a new these genes encode proteins, which are new cornified envelope gene cluster in the epidermal differentiation complex at human constituents distinct from SPRRs. Like SPRRs, the genes are 1q21 encoding a family of 18 proteins that are substrates for differentially expressed in different types of barrier epithelia. -

The Human Involucrin Gene Contains Spatially Distinct Regulatory Elements That Regulate Expression During Early Versus Late Epidermal DiErentiation
Oncogene (2002) 21, 738 ± 747 ã 2002 Nature Publishing Group All rights reserved 0950 ± 9232/02 $25.00 www.nature.com/onc The human involucrin gene contains spatially distinct regulatory elements that regulate expression during early versus late epidermal dierentiation James F Crish1, Frederic Bone1, Eric B Banks1 and Richard L Eckert*,1,2,3,4,5 1Department of Physiology and Biophysics, Case Western Reserve University School of Medicine, 2109 Adelbert Road, Cleveland, Ohio, OH 44106-4970, USA; 2Department of Dermatology, Case Western Reserve University School of Medicine, 2109 Adelbert Road, Cleveland, Ohio, OH 44106-4970, USA; 3Department of Reproductive Biology, Case Western Reserve University School of Medicine, 2109 Adelbert Road, Cleveland, Ohio, OH 44106-4970, USA; 4Department of Biochemistry, Case Western Reserve University School of Medicine, 2109 Adelbert Road, Cleveland, Ohio, OH 44106-4970, USA; 5Department of Oncology, Case Western Reserve University School of Medicine, 2109 Adelbert Road, Cleveland, Ohio, OH 44106-4970, USA Human involucrin (hINV) is a keratinocyte protein that Keywords: transgenic mice; activator protein-1; AP1; is expressed in the suprabasal compartment of the jun; fos; Sp1; gene regulation; epidermal keratinocyte; epidermis and other stratifying surface epithelia. In- keratinocyte dierentiation; tissue speci®c volucrin gene expression is initiated early in the dierentiation process and is maintained until terminal cell death. The distal regulatory region (DRR) is a Introduction segment of the hINV promoter (nucleotides 72473/ 71953) that accurately recapitulates the normal pattern The major cell type, and the cell type responsible for of suprabasal (spinous and granular layer) expression in the morphological characteristics of the epidermis, is transgenic mouse epithelia. -

Characterization of the Human Involucrin Promoter Using a Transient -Galactosidase Assay
Journal of Cell Science 103, 925-930 (1992) 925 Printed in Great Britain © The Company of Biologists Limited 1992 Characterization of the human involucrin promoter using a transient -galactosidase assay JOSEPH M. CARROLL1 and LORNE B. TAICHMAN2,* 1Graduate Program in Cellular and Developmental Biology, 2Department of Oral Biology and Pathology, School of Dental Medicine, State University of New York at Stony Brook, Stony Brook, New York 11794-8702, USA *Author for correspondence Summary Involucrin, a component of the cornified cell envelope, nascent RNA and suggested that sequences within the is expressed specifically in differentiating keratinocytes intron have regulatory activity. These results suggest of stratified squamous epithelia. To explore the regula- that the involucrin intron operates in vivo to regulate tion of involucrin expression, 3.7 kb of upstream expression in the epidermis. sequences of the human involucrin gene was cloned into a plasmid containing a -galactosidase reporter gene and transfected into early passage keratinocytes and a Abbreviations used: ADH, alcohol dehydrogenase; b-gal, b- galactosidase; DME, Dulbecco’s modified Eagle’s medium; variety of human cell types. The full-length construct DDAB, dimethyldioctyldecylammonium bromide; FCS, fetal calf gave maximal and tissue-specific expression. Deletion serum; kb, kilobase; ONPG, O-nitrophenyl b-D- analysis showed that sequences between 900 and 2500 galactopyranoside; PBS, phosphate buffered saline without bp upstream of the transcriptional start site and the calcium/magnesium salts; PCR, polymerase chain reaction; intron located between the transcriptional and transla- PtdEtn, dioleoyl-L-a -phosphatidylethanolamine; RSV, Rous tional start sites were required for maximal expression. sarcoma virus; SV40, simian virus 40. -

Involucrin - Structure and Role in Envelope Assembly
REVIEW Involucrin - Structure and Role in Envelope Assembly Richard L. Eckert, Michael B. Yaffe,james F. Crish, Shubha Murthy, Ellen A. Rorke, and Jean F. Welter Departments of Physiology and Biophysics (RLE, MBY, JFC, SM, EAR, JFW), Dermatology (RLE), Reproductive Biology (RLE), Biochemistry (RLE), and EnvI ronmental Health SCIences (EAR) , Case Western Reserve UniverSIty School of MedIcine, Cleveland, Ohio, U .S.A. he epidermis is elegantly designed to provide a con 'from cultured keratinocytes or fibroblasts [23] . This transfer is cal stantly renewing protective surface. The proliferating cium dependent and inhibited by TG inhibitors. Human involucrin stem cel ls, which constantly replenish the epidermis, has also been expressed in cultured rat keratinecytes, CHO cells, occupy the basal layer adjacent the basal lamina [1,2]. and PtK2 cells via vector-mediated gene transfer [24]. Elevation of The supra basal spinous and granular la yers contain intracellular calcium resulted in the disappearance of human inve T11s that have largely lost proliferative potential, but are still living lucrin frem the soluble phase in cells expressing keratinecyte (rat ce d functioning. These cells are undergoing a progressive process of ~eratinocytes) and tissue-type (CHO cell s) transglutaminase, respec ~ntrac ellular remodeling in preparation for terminal differentiation. tIvely [24]. In each case the disappea rance was inhibitable by cal r the stratum lucidum the remodeling is largely completed as the cium chelators or TG inhibitors. No disappearance from the soluble : ratinocytes pass from the living to the non-living state to form phase was observed in PtK2 cells, which express barely detectab le e neocytes [1,2]. -

University of California, San Diego
UC San Diego UC San Diego Electronic Theses and Dissertations Title The post-terminal differentiation fate of RNAs revealed by next-generation sequencing Permalink https://escholarship.org/uc/item/7324r1rj Author Lefkowitz, Gloria Kuo Publication Date 2012 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA, SAN DIEGO The post-terminal differentiation fate of RNAs revealed by next-generation sequencing A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Biomedical Sciences by Gloria Kuo Lefkowitz Committee in Charge: Professor Benjamin D. Yu, Chair Professor Richard Gallo Professor Bruce A. Hamilton Professor Miles F. Wilkinson Professor Eugene Yeo 2012 Copyright Gloria Kuo Lefkowitz, 2012 All rights reserved. The Dissertation of Gloria Kuo Lefkowitz is approved, and it is acceptable in quality and form for publication on microfilm and electronically: __________________________________________________________________ __________________________________________________________________ __________________________________________________________________ __________________________________________________________________ __________________________________________________________________ Chair University of California, San Diego 2012 iii DEDICATION Ma and Ba, for your early indulgence and support. Matt and James, for choosing more practical callings. Roy, my love, for patiently sharing the ups and downs -

Evaluation on Reprogramed Biological Processes in Transgenic Maize
www.nature.com/scientificreports OPEN Evaluation on reprogramed biological processes in transgenic maize varieties using transcriptomics and metabolomics Wei Fu1, Pengyu Zhu1, Mingnan Qu2, Wang Zhi1, Yongjiang Zhang1, Feiwu Li3* & Shuifang Zhu1* Genetic engineering (GM) has great potential to improve maize productivity, but rises some concerns on unintended efects, and equivalent as their comparators. There are some limitations through targeted analysis to detect the UE in genetically modifed organisms in many previous studies. We here reported a case-study on the efects of introducing herbicides and insect resistance (HIR) gene cassette on molecular profling (transcripts and metabolites) in a popular maize variety Zhengdan958 (ZD958) in China. We found that introducing HIR gene cassette bring a limited numbers of diferential abundant genes (DAGs) or diferential abundant metabolites (DAMs) between transgenic events and non-transgenic control. In contrast, averaged 10 times more DAGs and DAMs were observed when performed comparison under diferent growing environments in three diferent ecological regions of China than the numbers induced by gene efects. Major biological pathways relating to stress response or signaling transduction could explain somehow the efects of growing environments. We further compared two transgenic events mediated ZD958 (GM-ZD958) with either transgenic parent GM-Z58, and other genetic background nonGM-Z58, nonGM-ZD958, and Chang7-2. We found that the numbers of DAGs and DAMs between GM-ZD958 and its one parent maize variety, Z58 or GM-Z58 is equivalent, but not Chang7-2. These fndings suggest that greater efects due to diferent genetic background on altered molecular profling than gene modifcation itself. This study provides a case evidence indicating marginal efects of gene pleiotropic efects, and environmental efects should be emphasized. -

Codon Reiteration and the Evolution of Proteins
Proc. Nati. Acad. Sci. USA Vol. 91, pp. 4298-4302, May 1994 Evolution Codon reiteration and the evolution of proteins HOWARD GREEN AND NORMAN WANG Department of Cell Biology, Harvard Medical School, 25 Shattuck Street, Boston, MA 02115 Contributed by Howard Green, January 18, 1994 ABSTRACT Sequence data banks have been searched for dropped out, leaving few reiterants of hydrophobic amino proteins possessing uninterrupted reiterations of any amino acids. There is no reiterant of arginine. acid. Hydrophilic amino acids, and particularly glutamine, Among reiterants containing 20 or more residues, gluta- account for a large proportion of the longer reiterants. In the mine is clearly dominant and accounts for half of all reiter- genes for these proteins, the most common reiterants are those ants. The other nine amino acid residues found in reiterants that contain poly(CAG), even out-of-frame or, to a lesser are mainly hydrophilic. degree, those that contain repeated doublets ofCA, AG, or GC. The preferential generation of such reiterants requires that Codons Responsible for Reiterants of 10 or More Amino DNA strand-specific signals predispose to reiteration and thus Acid Residues to the extension of coding regions. Of the 229 reiterants, there are only 48 that are encoded by Sequences consisting of multiple consecutive residues of a uninterrupted reiterations of a single codon; most of the single amino acid (reiterants) are not rare in proteins. For amino acid reiterants are the result of mixed synonymous example, reiterants consisting of glutamine residues, most codons. Probably some of these were originally reiterants of often encoded by CAG, were first discovered in homeotic a single codon, but nucleotide substitutions have since oc- proteins (1-3) and later in other transcription factors. -

Cystatin-B Negatively Regulates the Malignant Characteristics of Oral Squamous Cell Carcinoma Possibly Via the Epithelium Proliferation/ Differentiation Program
ORIGINAL RESEARCH published: 24 August 2021 doi: 10.3389/fonc.2021.707066 Cystatin-B Negatively Regulates the Malignant Characteristics of Oral Squamous Cell Carcinoma Possibly Via the Epithelium Proliferation/ Differentiation Program Tian-Tian Xu 1, Xiao-Wen Zeng 1, Xin-Hong Wang 2, Lu-Xi Yang 1, Gang Luo 1* and Ting Yu 1* 1 Department of Periodontics, Affiliated Stomatology Hospital of Guangzhou Medical University, Guangzhou Key Laboratory of Basic and Applied Research of Oral Regenerative Medicine, Guangzhou, China, 2 Department of Oral Pathology and Medicine, Affiliated Stomatology Hospital of Guangzhou Medical University, Guangzhou Key Laboratory of Basic and Applied Research of Oral Regenerative Medicine, Guangzhou, China Disturbance in the proteolytic process is one of the malignant signs of tumors. Proteolysis Edited by: Eva Csosz, is highly orchestrated by cysteine cathepsin and its inhibitors. Cystatin-B (CSTB) is a University of Debrecen, Hungary general cysteine cathepsin inhibitor that prevents cysteine cathepsin from leaking from Reviewed by: lysosomes and causing inappropriate proteolysis. Our study found that CSTB was Csongor Kiss, downregulated in both oral squamous cell carcinoma (OSCC) tissues and cells University of Debrecen, Hungary Gergely Nagy, compared with normal controls. Immunohistochemical analysis showed that CSTB was University of Debrecen, Hungary mainly distributed in the epithelial structure of OSCC tissues, and its expression intensity *Correspondence: was related to the grade classification. A correlation analysis between CSTB and clinical Gang Luo [email protected] prognosis was performed using gene expression data and clinical information acquired Ting Yu from The Cancer Genome Atlas (TCGA) database. Patients with lower expression levels of [email protected] CSTB had shorter disease-free survival times and poorer clinicopathological features Specialty section: (e.g., lymph node metastases, perineural invasion, low degree of differentiation, and This article was submitted to advanced tumor stage).