Differentially Expressed Late Constituents of the Epidermal Cornified Envelope
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ONCOGENOMICS Through the Use of Chemotherapeutic Agents
Oncogene (2002) 21, 7749 – 7763 ª 2002 Nature Publishing Group All rights reserved 0950 – 9232/02 $25.00 www.nature.com/onc Expression profiling of primary non-small cell lung cancer for target identification Jim Heighway*,1, Teresa Knapp1, Lenetta Boyce1, Shelley Brennand1, John K Field1, Daniel C Betticher2, Daniel Ratschiller2, Mathias Gugger2, Michael Donovan3, Amy Lasek3 and Paula Rickert*,3 1Target Identification Group, Roy Castle International Centre for Lung Cancer Research, University of Liverpool, 200 London Road, Liverpool L3 9TA, UK; 2Institute of Medical Oncology, University of Bern, 3010 Bern, Switzerland; 3Incyte Genomics Inc., 3160 Porter Drive, Palo Alto, California, CA 94304, USA Using a panel of cDNA microarrays comprising 47 650 Introduction transcript elements, we have carried out a dual-channel analysis of gene expression in 39 resected primary The lack of generally effective therapies for lung cancer human non-small cell lung tumours versus normal lung leads to the high mortality rate associated with this tissue. Whilst *11 000 elements were scored as common disease. Recorded 5-year survival figures vary, differentially expressed at least twofold in at least one to some extent perhaps reflecting differences in sample, 96 transcripts were scored as over-represented ascertainment methods, but across Europe are approxi- fourfold or more in at least seven out of 39 tumours mately 10% (Bray et al., 2002). Whilst surgery is an and 30 sequences 16-fold in at least two out of 39 effective strategy for the treatment of localized lesions, tumours, including 24 transcripts in common. Tran- most patients present with overtly or covertly dissemi- scripts (178) were found under-represented fourfold in nated disease. -
Critical Evaluation of Gene Expression Changes in Human Tissues In
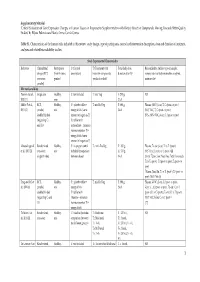
Supplementary Material ‘Critical Evaluation of Gene Expression Changes in Human Tissues in Response to Supplementation with Dietary Bioactive Compounds: Moving Towards Better-Quality Studies’ by Biljana Pokimica and María-Teresa García-Conesa Table S1. Characteristics of the human trials included in this review: study design, type of participants, control and intervention description, dose and duration of treatment, analyses and related bioavailability studies. Study Experimental Characteristics Reference Clinical trial Participants C (Control T (Treatment with Total daily dose, Bioavailability studies: type of sample, design (RCT, (health status, description) bioactive compounds, duration (d or h)1 compounds and (or) metabolites analysed, crossover, gender) products or diet) main results2 parallel) Mix meals and diets Persson I et al., Single arm Healthy, C: not included T: mix Veg T: 250 g, NR 2000 [1] men 21 d Møller P et al., RCT, Healthy, C1: placebo tablet + T: mix FruVeg T: 600 g, Plasma: (NS↑) β-car, T, C2 (post- vs pre-) 2003 [2] parallel, mix energy drink (same 24 d (NC) VitC, T, C2 (post- vs pre-) double blinded amount of sugars as T) (NS↓, 69%) VitC, β-car, C1 (post- vs pre-) (regarding C1 C2: tablet with and C2) antioxidants + minerals (same amount as T) + energy drink (same amount of sugars as T) Almendingen K Randomized, Healthy, C: no proper control T1,2: mix FruVeg T1: 300 g, Plasma: ↑α-car, β-car, T2 vs T1 (post-) et al., 2005 [3] crossover, mix included (comparison T2: 750 g, (NS↑) Lyc, Lut, T2 vs T1 (post-) [4] single -
Salivary Exrna Biomarkers to Detect Gingivitis and Monitor Disease Regression
Received: 3 December 2017 | Revised: 14 April 2018 | Accepted: 13 May 2018 DOI: 10.1111/jcpe.12930 OTHER Salivary exRNA biomarkers to detect gingivitis and monitor disease regression Karolina E. Kaczor-Urbanowicz1,2* | Harsh M. Trivedi3* | Patricia O. Lima1,4 | Paulo M. Camargo5 | William V. Giannobile6 | Tristan R. Grogan7 | Frederico O. Gleber-Netto8 | Yair Whiteman9 | Feng Li1 | Hyo Jung Lee10 | Karan Dharia11 | Katri Aro1 | Carmen Martin Carerras-Presas12 | Saarah Amuthan11 | Manjiri Vartak11 | David Akin1 | Hiba Al-adbullah11 | Kanika Bembey11 | Perry R. Klokkevold5 | David Elashoff7 | Virginia M. Barnes13 | Rose Richter13 | William DeVizio13 | James G. Masters3 | David T. W. Wong1 1UCLA School of Dentistry, Center for Oral/Head & Neck Oncology Research, University of California at Los Angeles, Los Angeles, California 2Section of Orthodontics, UCLA School of Dentistry, University of California at Los Angeles, Los Angeles, California 3Early Research Oral Care, Colgate Palmolive Co., Piscataway, New Jersey 4Department of Physiological Sciences, Piracicaba Dental School, University of Campinas, Piracicaba, São Paulo, Brazil 5Section of Periodontics, UCLA School of Dentistry, University of California at Los Angeles, Los Angeles, California 6Department of Periodontics and Oral Medicine, School of Dentistry, University of Michigan, Ann Arbor, Michigan 7Department of Biostatistics, University of California at Los Angeles, Los Angeles, California 8Medical Genomics Laboratory, Centro Internacional de Pesquisa e Ensino (CIPE), AC Camargo Cancer -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

Microarray Analysis of Novel Genes Involved in HSV- 2 Infection
Microarray analysis of novel genes involved in HSV- 2 infection Hao Zhang Nanjing University of Chinese Medicine Tao Liu ( [email protected] ) Nanjing University of Chinese Medicine https://orcid.org/0000-0002-7654-2995 Research Article Keywords: HSV-2 infection,Microarray analysis,Histospecic gene expression Posted Date: May 12th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-517057/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/19 Abstract Background: Herpes simplex virus type 2 infects the body and becomes an incurable and recurring disease. The pathogenesis of HSV-2 infection is not completely clear. Methods: We analyze the GSE18527 dataset in the GEO database in this paper to obtain distinctively displayed genes(DDGs)in the total sequential RNA of the biopsies of normal and lesioned skin groups, healed skin and lesioned skin groups of genital herpes patients, respectively.The related data of 3 cases of normal skin group, 4 cases of lesioned group and 6 cases of healed group were analyzed.The histospecic gene analysis , functional enrichment and protein interaction network analysis of the differential genes were also performed, and the critical components were selected. Results: 40 up-regulated genes and 43 down-regulated genes were isolated by differential performance assay. Histospecic gene analysis of DDGs suggested that the most abundant system for gene expression was the skin, immune system and the nervous system.Through the construction of core gene combinations, protein interaction network analysis and selection of histospecic distribution genes, 17 associated genes were selected CXCL10,MX1,ISG15,IFIT1,IFIT3,IFIT2,OASL,ISG20,RSAD2,GBP1,IFI44L,DDX58,USP18,CXCL11,GBP5,GBP4 and CXCL9.The above genes are mainly located in the skin, immune system, nervous system and reproductive system. -

University of California, San Diego
UC San Diego UC San Diego Electronic Theses and Dissertations Title The post-terminal differentiation fate of RNAs revealed by next-generation sequencing Permalink https://escholarship.org/uc/item/7324r1rj Author Lefkowitz, Gloria Kuo Publication Date 2012 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA, SAN DIEGO The post-terminal differentiation fate of RNAs revealed by next-generation sequencing A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Biomedical Sciences by Gloria Kuo Lefkowitz Committee in Charge: Professor Benjamin D. Yu, Chair Professor Richard Gallo Professor Bruce A. Hamilton Professor Miles F. Wilkinson Professor Eugene Yeo 2012 Copyright Gloria Kuo Lefkowitz, 2012 All rights reserved. The Dissertation of Gloria Kuo Lefkowitz is approved, and it is acceptable in quality and form for publication on microfilm and electronically: __________________________________________________________________ __________________________________________________________________ __________________________________________________________________ __________________________________________________________________ __________________________________________________________________ Chair University of California, San Diego 2012 iii DEDICATION Ma and Ba, for your early indulgence and support. Matt and James, for choosing more practical callings. Roy, my love, for patiently sharing the ups and downs -

Evaluation on Reprogramed Biological Processes in Transgenic Maize
www.nature.com/scientificreports OPEN Evaluation on reprogramed biological processes in transgenic maize varieties using transcriptomics and metabolomics Wei Fu1, Pengyu Zhu1, Mingnan Qu2, Wang Zhi1, Yongjiang Zhang1, Feiwu Li3* & Shuifang Zhu1* Genetic engineering (GM) has great potential to improve maize productivity, but rises some concerns on unintended efects, and equivalent as their comparators. There are some limitations through targeted analysis to detect the UE in genetically modifed organisms in many previous studies. We here reported a case-study on the efects of introducing herbicides and insect resistance (HIR) gene cassette on molecular profling (transcripts and metabolites) in a popular maize variety Zhengdan958 (ZD958) in China. We found that introducing HIR gene cassette bring a limited numbers of diferential abundant genes (DAGs) or diferential abundant metabolites (DAMs) between transgenic events and non-transgenic control. In contrast, averaged 10 times more DAGs and DAMs were observed when performed comparison under diferent growing environments in three diferent ecological regions of China than the numbers induced by gene efects. Major biological pathways relating to stress response or signaling transduction could explain somehow the efects of growing environments. We further compared two transgenic events mediated ZD958 (GM-ZD958) with either transgenic parent GM-Z58, and other genetic background nonGM-Z58, nonGM-ZD958, and Chang7-2. We found that the numbers of DAGs and DAMs between GM-ZD958 and its one parent maize variety, Z58 or GM-Z58 is equivalent, but not Chang7-2. These fndings suggest that greater efects due to diferent genetic background on altered molecular profling than gene modifcation itself. This study provides a case evidence indicating marginal efects of gene pleiotropic efects, and environmental efects should be emphasized. -

Supplementary Data
Supplemental figures Supplemental figure 1: Tumor sample selection. A total of 98 thymic tumor specimens were stored in Memorial Sloan-Kettering Cancer Center tumor banks during the study period. 64 cases corresponded to previously untreated tumors, which were resected upfront after diagnosis. Adjuvant treatment was delivered in 7 patients (radiotherapy in 4 cases, cyclophosphamide- doxorubicin-vincristine (CAV) chemotherapy in 3 cases). 34 tumors were resected after induction treatment, consisting of chemotherapy in 16 patients (cyclophosphamide-doxorubicin- cisplatin (CAP) in 11 cases, cisplatin-etoposide (PE) in 3 cases, cisplatin-etoposide-ifosfamide (VIP) in 1 case, and cisplatin-docetaxel in 1 case), in radiotherapy (45 Gy) in 1 patient, and in sequential chemoradiation (CAP followed by a 45 Gy-radiotherapy) in 1 patient. Among these 34 patients, 6 received adjuvant radiotherapy. 1 Supplemental Figure 2: Amino acid alignments of KIT H697 in the human protein and related orthologs, using (A) the Homologene database (exons 14 and 15), and (B) the UCSC Genome Browser database (exon 14). Residue H697 is highlighted with red boxes. Both alignments indicate that residue H697 is highly conserved. 2 Supplemental Figure 3: Direct comparison of the genomic profiles of thymic squamous cell carcinomas (n=7) and lung primary squamous cell carcinomas (n=6). (A) Unsupervised clustering analysis. Gains are indicated in red, and losses in green, by genomic position along the 22 chromosomes. (B) Genomic profiles and recurrent copy number alterations in thymic carcinomas and lung squamous cell carcinomas. Gains are indicated in red, and losses in blue. 3 Supplemental Methods Mutational profiling The exonic regions of interest (NCBI Human Genome Build 36.1) were broken into amplicons of 500 bp or less, and specific primers were designed using Primer 3 (on the World Wide Web for general users and for biologist programmers (see Supplemental Table 2) [1]. -

Cystatin-B Negatively Regulates the Malignant Characteristics of Oral Squamous Cell Carcinoma Possibly Via the Epithelium Proliferation/ Differentiation Program
ORIGINAL RESEARCH published: 24 August 2021 doi: 10.3389/fonc.2021.707066 Cystatin-B Negatively Regulates the Malignant Characteristics of Oral Squamous Cell Carcinoma Possibly Via the Epithelium Proliferation/ Differentiation Program Tian-Tian Xu 1, Xiao-Wen Zeng 1, Xin-Hong Wang 2, Lu-Xi Yang 1, Gang Luo 1* and Ting Yu 1* 1 Department of Periodontics, Affiliated Stomatology Hospital of Guangzhou Medical University, Guangzhou Key Laboratory of Basic and Applied Research of Oral Regenerative Medicine, Guangzhou, China, 2 Department of Oral Pathology and Medicine, Affiliated Stomatology Hospital of Guangzhou Medical University, Guangzhou Key Laboratory of Basic and Applied Research of Oral Regenerative Medicine, Guangzhou, China Disturbance in the proteolytic process is one of the malignant signs of tumors. Proteolysis Edited by: Eva Csosz, is highly orchestrated by cysteine cathepsin and its inhibitors. Cystatin-B (CSTB) is a University of Debrecen, Hungary general cysteine cathepsin inhibitor that prevents cysteine cathepsin from leaking from Reviewed by: lysosomes and causing inappropriate proteolysis. Our study found that CSTB was Csongor Kiss, downregulated in both oral squamous cell carcinoma (OSCC) tissues and cells University of Debrecen, Hungary Gergely Nagy, compared with normal controls. Immunohistochemical analysis showed that CSTB was University of Debrecen, Hungary mainly distributed in the epithelial structure of OSCC tissues, and its expression intensity *Correspondence: was related to the grade classification. A correlation analysis between CSTB and clinical Gang Luo [email protected] prognosis was performed using gene expression data and clinical information acquired Ting Yu from The Cancer Genome Atlas (TCGA) database. Patients with lower expression levels of [email protected] CSTB had shorter disease-free survival times and poorer clinicopathological features Specialty section: (e.g., lymph node metastases, perineural invasion, low degree of differentiation, and This article was submitted to advanced tumor stage). -

Analysis of the Ultraviolet B Response in Primary Human Keratinocytes Using Oligonucleotide Microarrays
Analysis of the ultraviolet B response in primary human keratinocytes using oligonucleotide microarrays Angela Sesto*, Manuel Navarro*†, Frank Burslem‡, and Jose´L. Jorcano* *Department of Molecular and Cell Biology, Centro de Investigaciones Energe´ticas, Medioambientales y Tecnolo´gicas (CIEMAT), Avenida Complutense 22, 28040 Madrid, Spain; and ‡Pfizer Global Research and Development, Sandwich, Kent CT13 9NJ, United Kingdom Communicated by A. Garcia-Bellido, Autonomous University of Madrid, Madrid, Spain, December 17, 2001 (received for review October 19, 2001) UV radiation is the most important environmental skin aggressor, To understand the mechanisms of UV response, we sought to causing cancer and other problems. This paper reports the use of obtain the transcriptional profile of primary keratinocytes after oligonucleotide microarray technology to determine changes in UVB irradiation. Cultured human epidermal keratinocytes were gene expression in human keratinocytes after UVB treatment. treated with three UV doses, and samples were collected at Examination of the effects of different doses at different times different intervals. Oligonucleotide microarrays containing over after irradiation gave a global picture of the keratinocyte response 6,000 genes (HuGeneFL, Affymetrix, Santa Clara, CA) were to this type of insult. Five hundred thirty-nine regulated transcripts used to quantitatively assess changes in gene expression. The were found and organized into nine different clusters depending subset of regulated transcripts was classified into self-organizing on behavior patterns. Classification of these genes into 23 func- maps according to their different behaviors. Biological functions tional categories revealed that several biological processes are were assigned to these genes, which were then correlated with globally affected by UVB. -

LJELSR: a Strengthened Version of JELSR for Feature Selection and Clustering
Article LJELSR: A Strengthened Version of JELSR for Feature Selection and Clustering Sha-Sha Wu 1, Mi-Xiao Hou 1, Chun-Mei Feng 1,2 and Jin-Xing Liu 1,* 1 School of Information Science and Engineering, Qufu Normal University, Rizhao 276826, China; [email protected] (S.-S.W.); [email protected] (M.-X.H.); [email protected] (C.-M.F.) 2 Bio-Computing Research Center, Harbin Institute of Technology, Shenzhen 518055, China * Correspondence: [email protected]; Tel.: +086-633-3981-241 Received: 4 December 2018; Accepted: 7 February 2019; Published: 18 February 2019 Abstract: Feature selection and sample clustering play an important role in bioinformatics. Traditional feature selection methods separate sparse regression and embedding learning. Later, to effectively identify the significant features of the genomic data, Joint Embedding Learning and Sparse Regression (JELSR) is proposed. However, since there are many redundancy and noise values in genomic data, the sparseness of this method is far from enough. In this paper, we propose a strengthened version of JELSR by adding the L1-norm constraint on the regularization term based on a previous model, and call it LJELSR, to further improve the sparseness of the method. Then, we provide a new iterative algorithm to obtain the convergence solution. The experimental results show that our method achieves a state-of-the-art level both in identifying differentially expressed genes and sample clustering on different genomic data compared to previous methods. Additionally, the selected differentially expressed genes may be of great value in medical research. Keywords: differentially expressed genes; feature selection; L1-norm; sample clustering; sparse constraint 1. -

Viewer Tool ( 4
BRIEF COMMUNICATION www.jasn.org Comparison of Glomerular and Podocyte mRNA Profiles in Streptozotocin-Induced Diabetes † † † ‡ † Jia Fu,* Chengguo Wei, Kyung Lee, Weijia Zhang, Wu He, Peter Chuang, Zhihong Liu,* † and John Cijiang He § *National Clinical Research Center of Kidney Diseases, Jinling Hospital, Nanjing University School of Medicine, Nanjing, China; †Division of Nephrology, Department of Medicine, ‡Flow Cytometry Shared Resource Facility, Icahn School of Medicine at Mount Sinai, New York; §Renal Program, James J. Peters VA Medical Center at Bronx, New York. ABSTRACT Evaluating the mRNA profile of podocytes in the diabetic kidney may indicate STZ induction alone.10 In order to spe- genes involved in the pathogenesis of diabetic nephropathy. To determine if the cifically label and isolate podoctyes, we 2 2 podocyte-specific gene information contained in mRNA profiles of the whole crossed the eNOS / mice with the IRG glomerulus of the diabetic kidney accurately reflects gene expression in the isolated mice11 that ubiquitously express a red 2/2 podocytes, we crossed Nos3 IRG mice with podocin-rtTA and TetON-Cre mice fluorescent protein prior to Cre-mediated for enhanced green fluorescent protein labeling of podocytes before diabetic injury. recombination and an enhanced green Diabetes was induced by streptozotocin, and mRNA profiles of isolated glomeruli fluorescent protein (EGFP) following re- and sorted podocytes from diabetic and control mice were examined 10 weeks combination. These mice were further later. Expression of podocyte-specific markers in glomeruli was downregulated in bred with podocin-rtTA and TetON-Cre diabetic mice compared with controls. However, expression of these markers was (LC1) transgenic mice for inducible not altered in sorted podocytes from diabetic mice.