Genes Encoding Structural Proteins of Epidermal Cornification
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
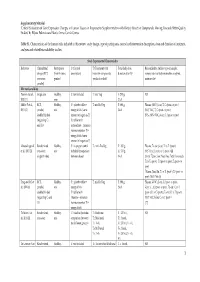
Critical Evaluation of Gene Expression Changes in Human Tissues In
Supplementary Material ‘Critical Evaluation of Gene Expression Changes in Human Tissues in Response to Supplementation with Dietary Bioactive Compounds: Moving Towards Better-Quality Studies’ by Biljana Pokimica and María-Teresa García-Conesa Table S1. Characteristics of the human trials included in this review: study design, type of participants, control and intervention description, dose and duration of treatment, analyses and related bioavailability studies. Study Experimental Characteristics Reference Clinical trial Participants C (Control T (Treatment with Total daily dose, Bioavailability studies: type of sample, design (RCT, (health status, description) bioactive compounds, duration (d or h)1 compounds and (or) metabolites analysed, crossover, gender) products or diet) main results2 parallel) Mix meals and diets Persson I et al., Single arm Healthy, C: not included T: mix Veg T: 250 g, NR 2000 [1] men 21 d Møller P et al., RCT, Healthy, C1: placebo tablet + T: mix FruVeg T: 600 g, Plasma: (NS↑) β-car, T, C2 (post- vs pre-) 2003 [2] parallel, mix energy drink (same 24 d (NC) VitC, T, C2 (post- vs pre-) double blinded amount of sugars as T) (NS↓, 69%) VitC, β-car, C1 (post- vs pre-) (regarding C1 C2: tablet with and C2) antioxidants + minerals (same amount as T) + energy drink (same amount of sugars as T) Almendingen K Randomized, Healthy, C: no proper control T1,2: mix FruVeg T1: 300 g, Plasma: ↑α-car, β-car, T2 vs T1 (post-) et al., 2005 [3] crossover, mix included (comparison T2: 750 g, (NS↑) Lyc, Lut, T2 vs T1 (post-) [4] single -

Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino-Like Mouse Models: Tracking the Role of the Hairless Gene
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Doctoral Dissertations Graduate School 5-2006 Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino-like Mouse Models: Tracking the Role of the Hairless Gene Yutao Liu University of Tennessee - Knoxville Follow this and additional works at: https://trace.tennessee.edu/utk_graddiss Part of the Life Sciences Commons Recommended Citation Liu, Yutao, "Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino- like Mouse Models: Tracking the Role of the Hairless Gene. " PhD diss., University of Tennessee, 2006. https://trace.tennessee.edu/utk_graddiss/1824 This Dissertation is brought to you for free and open access by the Graduate School at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Doctoral Dissertations by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. To the Graduate Council: I am submitting herewith a dissertation written by Yutao Liu entitled "Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino-like Mouse Models: Tracking the Role of the Hairless Gene." I have examined the final electronic copy of this dissertation for form and content and recommend that it be accepted in partial fulfillment of the requirements for the degree of Doctor of Philosophy, with a major in Life Sciences. Brynn H. Voy, Major Professor We have read this dissertation and recommend its acceptance: Naima Moustaid-Moussa, Yisong Wang, Rogert Hettich Accepted for the Council: Carolyn R. -

Differential Roles of P63 Isoforms in Epidermal Development: Selective Genetic Complementation in P63 Null Mice
Cell Death and Differentiation (2006) 13, 1037–1047 & 2006 Nature Publishing Group All rights reserved 1350-9047/06 $30.00 www.nature.com/cdd Differential roles of p63 isoforms in epidermal development: selective genetic complementation in p63 null mice E Candi1, A Rufini1, A Terrinoni1, D Dinsdale2, M Ranalli1, The skin consists of two compartments, the dermis and the A Paradisi1, V De Laurenzi2, LG Spagnoli1, MV Catani1, epidermis. The latter is a multilayered, stratified epithelium S Ramadan1, RA Knight2 and G Melino*,1,2 continuously regenerated by terminally differentiating keratino- cytes, a process called cornification1–3 (Figure 1a and b). 4 1 Biochemistry Laboratory, IDI-IRCCS, c/o University of Rome ‘Tor Vergata’, Recent evidence demonstrates a major role for p63, a 00133 Rome, Italy member of the p53 family,5–8 in this process as mutations in 2 Medical Research Council, Toxicology Unit, Leicester University, Leicester the TP63 gene cause limb and skin defects in humans,9 and LE1 9HN, UK p63À/À mice have no epidermis, no limbs and die at birth * Corresponding author: G Melino, Medical Research Council, Toxicology Unit, owing to dehydration.10,11 Hodgkin Building, Leicester University, Lancaster Road, PO Box 138, Leicester LE1 9HN, UK. Tel: þ 44 116 252 5616; Fax: þ 44 116 252 5551; The expression of p63 proteins originates from two E-mail: [email protected] promoters, giving rise to TAp63 and DNp63 isoforms. In addition, both isoforms undergo alternative splicing at the Received 03.3.06; revised 08.3.06; accepted 08.3.06; published online 07.4.06 C-terminus producing three different TAp63 and DNp63 Edited by P Vandenabeele isoforms (a, b and g). -

Downloaded from Genomic Data Common Website (GDC at Accessed on 2019)
G C A T T A C G G C A T genes Article Molecular Pathways Associated with Kallikrein 6 Overexpression in Colorectal Cancer Ritu Pandey 1,2,*, Muhan Zhou 3, Yuliang Chen 3, Dalila Darmoul 4 , Conner C. Kisiel 2, Valentine N. Nfonsam 5 and Natalia A. Ignatenko 1,2 1 Department of Cellular and Molecular Medicine, University of Arizona, Tucson, AZ 85721, USA; [email protected] 2 University of Arizona Cancer Center, University of Arizona, Tucson, AZ 85724, USA; [email protected] 3 Bioinformatics Shared Resource, University of Arizona Cancer Center, Tucson, AZ 85724, USA; [email protected] (M.Z.); [email protected] (Y.C.) 4 Institut National de la Santé et de la Recherche Médicale (INSERM), Université de Paris, Lariboisière Hospital, 75010 Paris, France; [email protected] 5 Department of Surgery, Section of Surgical Oncology, University of Arizona, Tucson, AZ 85724, USA; [email protected] * Correspondence: [email protected] Abstract: Colorectal cancer (CRC) remains one of the leading causes of cancer-related death world- wide. The high mortality of CRC is related to its ability to metastasize to distant organs. The kallikrein-related peptidase Kallikrein 6 (KLK6) is overexpressed in CRC and contributes to cancer cell invasion and metastasis. The goal of this study was to identify KLK6-associated markers for the CRC prognosis and treatment. Tumor Samples from the CRC patients with significantly elevated Citation: Pandey, R.; Zhou, M.; Chen, KLK6 transcript levels were identified in the RNA-Seq data from Cancer Genome Atlas (TCGA) Y.; Darmoul, D.; Kisiel, C.C.; and their expression profiles were evaluated using Gene Ontology (GO), Phenotype and Reactome Nfonsam, V.N.; Ignatenko, N.A. -

Cell Shape Controls Terminal Differentiation of Human Epidermal Keratinocytes (Cell Adhesion/Proliferation/Involucrin Expression) FIONA M
Proc. Natl. Acad. Sci. USA Vol. 85, pp. 5576-5580, August 1988 Cell Biology Cell shape controls terminal differentiation of human epidermal keratinocytes (cell adhesion/proliferation/involucrin expression) FIONA M. WATT*, PETER W. JORDANt, AND CHARLES H. O'NEILLt *Keratinocyte Laboratory, and tAnchorage Laboratory, Imperial Cancer Research Fund, P.O. Box 123, Lincoln's Inn Fields, London WC2A 3PX, United Kingdom Communicated by Howard Green, May 2, 1988 ABSTRACT Cultures of human epidermal keratinocytes To investigate the role of cell-substratum contact, and provide a useful experimental model with which to study the hence cell shape, in regulating terminal differentiation, we factors that regulate cell proliferation and terminal differen- have made use of a recently described technique that allows tiation. One situation that is known to trigger premature precise control of cell-substratum contact in the absence of terminal differentiation is suspension culture, when keratin- cell-cell contact (14). We have looked at the effect ofchanges ocytes are deprived of substratum and intercellular contact. in cell shape on DNA synthesis and involucrin expression We have now investigated whether area ofsubstratum contact, and conclude that a rounded morphology acts as a signal to and hence cell shape, can regulate terminal differentiation. keratinocytes to stop dividing and to undergo terminal Keratinocytes were grown on circular adhesive islands that differentiation; inhibition ofproliferation in spread cells is not prevented cell-cell contact. By varying island area we could a sufficient signal for terminal differentiation. vary cell shape from fully spread to almost spherical. We found that when substratum contact was restricted, DNA synthesis was inhibited and expression of involucrin, a marker of MATERIALS AND METHODS terminal differentiation, was stimulated. -

Microarray Analysis of Novel Genes Involved in HSV- 2 Infection
Microarray analysis of novel genes involved in HSV- 2 infection Hao Zhang Nanjing University of Chinese Medicine Tao Liu ( [email protected] ) Nanjing University of Chinese Medicine https://orcid.org/0000-0002-7654-2995 Research Article Keywords: HSV-2 infection,Microarray analysis,Histospecic gene expression Posted Date: May 12th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-517057/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/19 Abstract Background: Herpes simplex virus type 2 infects the body and becomes an incurable and recurring disease. The pathogenesis of HSV-2 infection is not completely clear. Methods: We analyze the GSE18527 dataset in the GEO database in this paper to obtain distinctively displayed genes(DDGs)in the total sequential RNA of the biopsies of normal and lesioned skin groups, healed skin and lesioned skin groups of genital herpes patients, respectively.The related data of 3 cases of normal skin group, 4 cases of lesioned group and 6 cases of healed group were analyzed.The histospecic gene analysis , functional enrichment and protein interaction network analysis of the differential genes were also performed, and the critical components were selected. Results: 40 up-regulated genes and 43 down-regulated genes were isolated by differential performance assay. Histospecic gene analysis of DDGs suggested that the most abundant system for gene expression was the skin, immune system and the nervous system.Through the construction of core gene combinations, protein interaction network analysis and selection of histospecic distribution genes, 17 associated genes were selected CXCL10,MX1,ISG15,IFIT1,IFIT3,IFIT2,OASL,ISG20,RSAD2,GBP1,IFI44L,DDX58,USP18,CXCL11,GBP5,GBP4 and CXCL9.The above genes are mainly located in the skin, immune system, nervous system and reproductive system. -

High-Throughput Bioinformatics Approaches to Understand Gene Expression Regulation in Head and Neck Tumors
High-throughput bioinformatics approaches to understand gene expression regulation in head and neck tumors by Yanxiao Zhang A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Bioinformatics) in The University of Michigan 2016 Doctoral Committee: Associate Professor Maureen A. Sartor, Chair Professor Thomas E. Carey Assistant Professor Hui Jiang Professor Ronald J. Koenig Associate Professor Laura M. Rozek Professor Kerby A. Shedden c Yanxiao Zhang 2016 All Rights Reserved I dedicate this thesis to my family. For their unfailing love, understanding and support. ii ACKNOWLEDGEMENTS I would like to express my gratitude to Dr. Maureen Sartor for her guidance in my research and career development. She is a great mentor. She patiently taught me when I started new in this field, granted me freedom to explore and helped me out when I got lost. Her dedication to work, enthusiasm in teaching, mentoring and communicating science have inspired me to feel the excite- ment of research beyond novel scientific discoveries. I’m also grateful to have an interdisciplinary committee. Their feedback on my research progress and presentation skills is very valuable. In particular, I would like to thank Dr. Thomas Carey and Dr. Laura Rozek for insightful discussions on the biology of head and neck cancers and human papillomavirus, Dr. Ronald Koenig for expert knowledge on thyroid cancers, Dr. Hui Jiang and Dr. Kerby Shedden for feedback on the statistics part of my thesis. I would like to thank all the past and current members of Sartor lab for making the lab such a lovely place to stay and work in. -

Identification of Differentially Expressed Genes in Human Bladder Cancer Through Genome-Wide Gene Expression Profiling
521-531 24/7/06 18:28 Page 521 ONCOLOGY REPORTS 16: 521-531, 2006 521 Identification of differentially expressed genes in human bladder cancer through genome-wide gene expression profiling KAZUMORI KAWAKAMI1,3, HIDEKI ENOKIDA1, TOKUSHI TACHIWADA1, TAKENARI GOTANDA1, KENGO TSUNEYOSHI1, HIROYUKI KUBO1, KENRYU NISHIYAMA1, MASAKI TAKIGUCHI2, MASAYUKI NAKAGAWA1 and NAOHIKO SEKI3 1Department of Urology, Graduate School of Medical and Dental Sciences, Kagoshima University, 8-35-1 Sakuragaoka, Kagoshima 890-8520; Departments of 2Biochemistry and Genetics, and 3Functional Genomics, Graduate School of Medicine, Chiba University, 1-8-1 Inohana, Chuo-ku, Chiba 260-8670, Japan Received February 15, 2006; Accepted April 27, 2006 Abstract. Large-scale gene expression profiling is an effective CKS2 gene not only as a potential biomarker for diagnosing, strategy for understanding the progression of bladder cancer but also for staging human BC. This is the first report (BC). The aim of this study was to identify genes that are demonstrating that CKS2 expression is strongly correlated expressed differently in the course of BC progression and to with the progression of human BC. establish new biomarkers for BC. Specimens from 21 patients with pathologically confirmed superficial (n=10) or Introduction invasive (n=11) BC and 4 normal bladder samples were studied; samples from 14 of the 21 BC samples were subjected Bladder cancer (BC) is among the 5 most common to microarray analysis. The validity of the microarray results malignancies worldwide, and the 2nd most common tumor of was verified by real-time RT-PCR. Of the 136 up-regulated the genitourinary tract and the 2nd most common cause of genes we detected, 21 were present in all 14 BCs examined death in patients with cancer of the urinary tract (1-7). -

Differentially Expressed Late Constituents of the Epidermal Cornified Envelope
Differentially expressed late constituents of the epidermal cornified envelope D. Marshall, M. J. Hardman, K. M. Nield, and C. Byrne* School of Biological Sciences, University of Manchester, 3.239 Stopford Building, Oxford Road, Manchester M13 9PT, United Kingdom Communicated by Elaine Fuchs, University of Chicago, Chicago, IL, September 17, 2001 (received for review June 23, 2001) Barrier activity of skin and internal barrier-forming epithelial There have been persistent reports of additional genes͞ linings are conferred by a lipid-corneocyte structure (stratum proteins with structures homologous to cornified envelope pro- corneum in skin).The integrity of the corneocytes depends on the teins. These include XP5, XP31, XP32 (12), the newly identified outer cornified envelope and is essential for maintenance of barrier component of the EDC (NICE-1; ref. 13), protein products of a function. During epidermal development and differentiation, pro- range of annotated expressed sequence tags (ESTs; ref. 14), and teins are sequentially incorporated into the envelope via action of Eig3 protein (15). We show that at least some of these proteins epidermal transglutaminases in a well documented process. How- are encoded by a previously undetected gene cluster in the ever, recent knockouts of major cornified envelope constituents human epidermal differentiation complex (EDC) at 1q21 (16), have failed to disrupt barrier function significantly, suggesting that with homologues detected in mouse (12, 14, 16). We show that additional unidentified components are involved. We report a new these genes encode proteins, which are new cornified envelope gene cluster in the epidermal differentiation complex at human constituents distinct from SPRRs. Like SPRRs, the genes are 1q21 encoding a family of 18 proteins that are substrates for differentially expressed in different types of barrier epithelia. -

The Human Involucrin Gene Contains Spatially Distinct Regulatory Elements That Regulate Expression During Early Versus Late Epidermal DiErentiation
Oncogene (2002) 21, 738 ± 747 ã 2002 Nature Publishing Group All rights reserved 0950 ± 9232/02 $25.00 www.nature.com/onc The human involucrin gene contains spatially distinct regulatory elements that regulate expression during early versus late epidermal dierentiation James F Crish1, Frederic Bone1, Eric B Banks1 and Richard L Eckert*,1,2,3,4,5 1Department of Physiology and Biophysics, Case Western Reserve University School of Medicine, 2109 Adelbert Road, Cleveland, Ohio, OH 44106-4970, USA; 2Department of Dermatology, Case Western Reserve University School of Medicine, 2109 Adelbert Road, Cleveland, Ohio, OH 44106-4970, USA; 3Department of Reproductive Biology, Case Western Reserve University School of Medicine, 2109 Adelbert Road, Cleveland, Ohio, OH 44106-4970, USA; 4Department of Biochemistry, Case Western Reserve University School of Medicine, 2109 Adelbert Road, Cleveland, Ohio, OH 44106-4970, USA; 5Department of Oncology, Case Western Reserve University School of Medicine, 2109 Adelbert Road, Cleveland, Ohio, OH 44106-4970, USA Human involucrin (hINV) is a keratinocyte protein that Keywords: transgenic mice; activator protein-1; AP1; is expressed in the suprabasal compartment of the jun; fos; Sp1; gene regulation; epidermal keratinocyte; epidermis and other stratifying surface epithelia. In- keratinocyte dierentiation; tissue speci®c volucrin gene expression is initiated early in the dierentiation process and is maintained until terminal cell death. The distal regulatory region (DRR) is a Introduction segment of the hINV promoter (nucleotides 72473/ 71953) that accurately recapitulates the normal pattern The major cell type, and the cell type responsible for of suprabasal (spinous and granular layer) expression in the morphological characteristics of the epidermis, is transgenic mouse epithelia. -

Characterization of the Human Involucrin Promoter Using a Transient -Galactosidase Assay
Journal of Cell Science 103, 925-930 (1992) 925 Printed in Great Britain © The Company of Biologists Limited 1992 Characterization of the human involucrin promoter using a transient -galactosidase assay JOSEPH M. CARROLL1 and LORNE B. TAICHMAN2,* 1Graduate Program in Cellular and Developmental Biology, 2Department of Oral Biology and Pathology, School of Dental Medicine, State University of New York at Stony Brook, Stony Brook, New York 11794-8702, USA *Author for correspondence Summary Involucrin, a component of the cornified cell envelope, nascent RNA and suggested that sequences within the is expressed specifically in differentiating keratinocytes intron have regulatory activity. These results suggest of stratified squamous epithelia. To explore the regula- that the involucrin intron operates in vivo to regulate tion of involucrin expression, 3.7 kb of upstream expression in the epidermis. sequences of the human involucrin gene was cloned into a plasmid containing a -galactosidase reporter gene and transfected into early passage keratinocytes and a Abbreviations used: ADH, alcohol dehydrogenase; b-gal, b- galactosidase; DME, Dulbecco’s modified Eagle’s medium; variety of human cell types. The full-length construct DDAB, dimethyldioctyldecylammonium bromide; FCS, fetal calf gave maximal and tissue-specific expression. Deletion serum; kb, kilobase; ONPG, O-nitrophenyl b-D- analysis showed that sequences between 900 and 2500 galactopyranoside; PBS, phosphate buffered saline without bp upstream of the transcriptional start site and the calcium/magnesium salts; PCR, polymerase chain reaction; intron located between the transcriptional and transla- PtdEtn, dioleoyl-L-a -phosphatidylethanolamine; RSV, Rous tional start sites were required for maximal expression. sarcoma virus; SV40, simian virus 40. -

Involucrin - Structure and Role in Envelope Assembly
REVIEW Involucrin - Structure and Role in Envelope Assembly Richard L. Eckert, Michael B. Yaffe,james F. Crish, Shubha Murthy, Ellen A. Rorke, and Jean F. Welter Departments of Physiology and Biophysics (RLE, MBY, JFC, SM, EAR, JFW), Dermatology (RLE), Reproductive Biology (RLE), Biochemistry (RLE), and EnvI ronmental Health SCIences (EAR) , Case Western Reserve UniverSIty School of MedIcine, Cleveland, Ohio, U .S.A. he epidermis is elegantly designed to provide a con 'from cultured keratinocytes or fibroblasts [23] . This transfer is cal stantly renewing protective surface. The proliferating cium dependent and inhibited by TG inhibitors. Human involucrin stem cel ls, which constantly replenish the epidermis, has also been expressed in cultured rat keratinecytes, CHO cells, occupy the basal layer adjacent the basal lamina [1,2]. and PtK2 cells via vector-mediated gene transfer [24]. Elevation of The supra basal spinous and granular la yers contain intracellular calcium resulted in the disappearance of human inve T11s that have largely lost proliferative potential, but are still living lucrin frem the soluble phase in cells expressing keratinecyte (rat ce d functioning. These cells are undergoing a progressive process of ~eratinocytes) and tissue-type (CHO cell s) transglutaminase, respec ~ntrac ellular remodeling in preparation for terminal differentiation. tIvely [24]. In each case the disappea rance was inhibitable by cal r the stratum lucidum the remodeling is largely completed as the cium chelators or TG inhibitors. No disappearance from the soluble : ratinocytes pass from the living to the non-living state to form phase was observed in PtK2 cells, which express barely detectab le e neocytes [1,2].