Differ in a Single Amino Acid-Asparagine to Aspartic Acid
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Amino Acid Chemistry
Handout 4 Amino Acid and Protein Chemistry ANSC 619 PHYSIOLOGICAL CHEMISTRY OF LIVESTOCK SPECIES Amino Acid Chemistry I. Chemistry of amino acids A. General amino acid structure + HN3- 1. All amino acids are carboxylic acids, i.e., they have a –COOH group at the #1 carbon. 2. All amino acids contain an amino group at the #2 carbon (may amino acids have a second amino group). 3. All amino acids are zwitterions – they contain both positive and negative charges at physiological pH. II. Essential and nonessential amino acids A. Nonessential amino acids: can make the carbon skeleton 1. From glycolysis. 2. From the TCA cycle. B. Nonessential if it can be made from an essential amino acid. 1. Amino acid "sparing". 2. May still be essential under some conditions. C. Essential amino acids 1. Branched chain amino acids (isoleucine, leucine and valine) 2. Lysine 3. Methionine 4. Phenyalanine 5. Threonine 6. Tryptophan 1 Handout 4 Amino Acid and Protein Chemistry D. Essential during rapid growth or for optimal health 1. Arginine 2. Histidine E. Nonessential amino acids 1. Alanine (from pyruvate) 2. Aspartate, asparagine (from oxaloacetate) 3. Cysteine (from serine and methionine) 4. Glutamate, glutamine (from α-ketoglutarate) 5. Glycine (from serine) 6. Proline (from glutamate) 7. Serine (from 3-phosphoglycerate) 8. Tyrosine (from phenylalanine) E. Nonessential and not required for protein synthesis 1. Hydroxyproline (made postranslationally from proline) 2. Hydroxylysine (made postranslationally from lysine) III. Acidic, basic, polar, and hydrophobic amino acids A. Acidic amino acids: amino acids that can donate a hydrogen ion (proton) and thereby decrease pH in an aqueous solution 1. -

Nucleotide Base Coding and Am1ino Acid Replacemients in Proteins* by Emil L
VOL. 48, 1962 BIOCHEMISTRY: E. L. SAIITH 677 18 Britten, R. J., and R. B. Roberts, Science, 131, 32 (1960). '9 Crestfield, A. M., K. C. Smith, and F. WV. Allen, J. Biol. Chem., 216, 185 (1955). 20 Gamow, G., Nature, 173, 318 (1954). 21 Brenner, S., these PROCEEDINGS, 43, 687 (1957). 22 Nirenberg, M. WV., J. H. Matthaei, and 0. WV. Jones, unpublished data. 23 Crick, F. H. C., L. Barnett, S. Brenner, and R. J. Watts-Tobin, Nature, 192, 1227 (1961). 24 Levene, P. A., and R. S. Tipson, J. Biol. Ch-nn., 111, 313 (1935). 25 Gierer, A., and K. W. Mundry, Nature, 182, 1437 (1958). 2' Tsugita, A., and H. Fraenkel-Conrat, J. Mllot. Biol., in press. 27 Tsugita, A., and H. Fraenkel-Conrat, personal communication. 28 Wittmann, H. G., Naturwissenschaften, 48, 729 (1961). 29 Freese, E., in Structure and Function of Genetic Elements, Brookhaven Symposia in Biology, no. 12 (1959), p. 63. NUCLEOTIDE BASE CODING AND AM1INO ACID REPLACEMIENTS IN PROTEINS* BY EMIL L. SMITHt LABORATORY FOR STUDY OF HEREDITARY AND METABOLIC DISORDERS AND THE DEPARTMENTS OF BIOLOGICAL CHEMISTRY AND MEDICINE, UNIVERSITY OF UTAH COLLEGE OF MEDICINE Communicated by Severo Ochoa, February 14, 1962 The problem of which bases of messenger or template RNA' specify the coding of amino acids in proteins has been largely elucidated by the use of synthetic polyri- bonucleotides.2-7 For these triplet nucleotide compositions (Table 1), it is of in- terest to examine some of the presently known cases of amino acid substitutions in polypeptides or proteins of known structure. -

An Integrated Meta-Analysis of Peripheral Blood Metabolites and Biological Functions in Major Depressive Disorder
Molecular Psychiatry https://doi.org/10.1038/s41380-020-0645-4 ARTICLE An integrated meta-analysis of peripheral blood metabolites and biological functions in major depressive disorder 1,2,3 1,2,3 1,2,3 1,3 1,3 4,5 1,3 1,3 Juncai Pu ● Yiyun Liu ● Hanping Zhang ● Lu Tian ● Siwen Gui ● Yue Yu ● Xiang Chen ● Yue Chen ● 1,2,3 1,3 1,3 1,3 1,3 1,2,3 Lining Yang ● Yanqin Ran ● Xiaogang Zhong ● Shaohua Xu ● Xuemian Song ● Lanxiang Liu ● 1,2,3 1,3 1,2,3 Peng Zheng ● Haiyang Wang ● Peng Xie Received: 3 June 2019 / Revised: 24 December 2019 / Accepted: 10 January 2020 © The Author(s) 2020. This article is published with open access Abstract Major depressive disorder (MDD) is a serious mental illness, characterized by high morbidity, which has increased in recent decades. However, the molecular mechanisms underlying MDD remain unclear. Previous studies have identified altered metabolic profiles in peripheral tissues associated with MDD. Using curated metabolic characterization data from a large sample of MDD patients, we meta-analyzed the results of metabolites in peripheral blood. Pathway and network analyses were then performed to elucidate the biological themes within these altered metabolites. We identified 23 differentially 1234567890();,: 1234567890();,: expressed metabolites between MDD patients and controls from 46 studies. MDD patients were characterized by higher levels of asymmetric dimethylarginine, tyramine, 2-hydroxybutyric acid, phosphatidylcholine (32:1), and taurochenode- soxycholic acid and lower levels of L-acetylcarnitine, creatinine, L-asparagine, L-glutamine, linoleic acid, pyruvic acid, palmitoleic acid, L-serine, oleic acid, myo-inositol, dodecanoic acid, L-methionine, hypoxanthine, palmitic acid, L-tryptophan, kynurenic acid, taurine, and 25-hydroxyvitamin D compared with controls. -
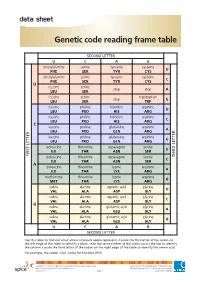
Genetic Code Reading Frame Table
data sheet Genetic code reading frame table SECOND LETTER U C A G phenylalanine serine tyrosine cysteine U PHE SER TYR CYS phenylalanine serine tyrosine cysteine C PHE SER TYR CYS U leucine serine stop stop A LEU SER leucine serine tryptophan stop G LEU SER TRP leucine proline histidine arginine U LEU PRO HIS ARG leucine proline histidine arginine C LEU PRO HIS ARG C leucine proline glutamine arginine A LEU PRO GLN ARG leucine proline glutamine arginine G LEU PRO GLN ARG isoleucine threonine asparagine serine U ILE THR ASN SER FIRST LETTER isoleucine threonine asparagine serine LETTERTHIRD C ILE THR ASN SER A isoleucine threonine lysine arginine A ILE THR LYS ARG methionine threonine lysine arginine G MET THR LYS ARG valine alanine aspartic acid glycine U VAL ALA ASP GLY valine alanine aspartic acid glycine C VAL ALA ASP GLY G valine alanine glutamic acid glycine A VAL ALA GLU GLY valine alanine glutamic acid glycine G VAL ALA GLU GLY U C A G SECOND LETTER Use this table to find out what amino acid each codon represents. Locate the first letter of the codon on the left edge of the table to identify a block, then the second letter of the codon across the top to identify the column. Locate the third letter of the codon on the right edge of the table to identify the amino acid. For example, the codon ‘CAU’ codes for histidine (HIS). ast0812 | Proteins 3: Genetic code reading frame table (data sheet) developed for the Department of Education WA © The University of Western Australia 2012 for conditions of use see spice.wa.edu.au/usage version 1.0 page 1 Licensed for NEALS. -

Analysis of Amino Acids by HPLC
Analysis of Amino Acids by HPLC Rita Steed Agilent Technologies, Inc. 800-227-9770 opt 3/opt3/opt 2 Amino Acid Analysis - Agilent Restricted Page 1 June 24, 2010 Outline • Amino Acids – Structure, Chemistry • Separation Considerations • Challenges • Instrumentation • Derivatization – OPA, FMOC • Overview of Separations • Examples Amino Acid Analysis - Agilent Restricted Page 2 June 24, 2010 Amino Acids – Structure, Chemistry CH3 Alanine (()Ala) Glutamic Acid (()Glu) Amino Acid Analysis - Agilent Restricted Page 3 June 24, 2010 Amino Acids – Zwitterionic Amino Acid Analysis - Agilent Restricted Page 4 June 24, 2010 Separation Considerations • Zwitterions - poor solubility near iso- …electric point • Most have poor UV absorbance • Derivatization – OPA, FMOC •Reduce polarity – increases retention in reversed-phase chromatoggpyraphy •Improve sensitivity – UV, Fluorescence • Detector; DAD, FLD, MS, ELSD Amino Acid Analysis - Agilent Restricted Page 5 June 24, 2010 Ortho Phthalaldehyde (OPA) and Fluorenylmethoxy chloroformate (FMOC) Reactions with Amines OPA O SR’ R’SH H NR +RNH2 H Room Temperature O Fluorescence: Ex 340nm, Em 450nm Non-fluorescent DAD: 338 , 10nm; Ref . 390 , 20nm Does not absorb at 338nm FMOC RR’NH - HCl + or Room Temperature RNH2 NRR’ or NHR Fluorescence: Ex 260nm, Em 325nm Fluorescent DAD: 262, 16nm; Ref. 324,8nm Absorbs at 262nm and Fluorescences at 324nm Group/Presentation Title Agilent Restricted Month ##, 200X Names and Order of Elution for OPA and FMOC Derivatives of Amino Acids Peak # AA Name AA Abbreviation Derivative Type Peak # AA Name AA Abbreviation Derivative Type Group/Presentation Title Agilent Restricted Month ##, 200X Agilent AAA Methods - They’ve Evolved • Automated Amino Acid Analysis – AminoQuant I & II (1987) •1090 •1100, Pub. -

Safety Assessment of Α-Amino Acids As Used in Cosmetics
Safety Assessment of α-Amino Acids as Used in Cosmetics Status: Final Report for Public Distribution Release Date: October 5, 2012 Panel Meeting Date: September 10-11, 2012 The 2012 Cosmetic Ingredient Review Expert Panel members are: Chairman, Wilma F. Bergfeld, M.D., F.A.C.P.; Donald V. Belsito, M.D.; Curtis D. Klaassen, Ph.D.; Daniel C. Liebler, Ph.D.; Ronald A. Hill, Ph.D. James G. Marks, Jr., M.D.; Ronald C. Shank, Ph.D.; Thomas J. Slaga, Ph.D.; and Paul W. Snyder, D.V.M., Ph.D. The CIR Director is F. Alan Andersen, Ph.D. This safety assessment was prepared by Christina L. Burnett, Scientific Analyst/Writer, and Bart Heldreth, Ph.D., Chemist CIR. © Cosmetic Ingredient Review 1101 17th Street, NW, Suite 412 Washington, DC 20036-4702 ph 202.331.0651 fax 202.331.0088 [email protected] 1 ABSTRACT The Cosmetic Ingredient Review Expert Panel (the Panel) reviewed the safety of α-amino acids, which function primarily as hair and skin conditioning agents in cosmetic products. The safety of α-amino acids as direct food additives has been well established based on extensive research through acute and chronic dietary exposures. The Panel focused its review on dermal irritation and sensitization data relevant to the use of these ingredients in topical cosmetics. The Panel concluded that α-amino acids were safe as cosmetic ingredients in the practices of use and concentration of this safety assessment. INTRODUCTION Amino acids and their salts are widely used as cosmetic ingredients, and function primarily as hair conditioning agents and skin conditioning agents (humectant and miscellaneous). -

THE CONDUCTIVITIES of AQUEOUS SOLUTIONS of GLYCINE, D,/-VALINE, and I-ASPARAGINE*
THE CONDUCTIVITIES OF AQUEOUS SOLUTIONS OF GLYCINE, d,/-VALINE, AND I-ASPARAGINE* B~ JOHN W. MEI-IL AND CARL L. A. SCHMIDT (From the Division of Biochemistry, Unirersity of California Medical School, Berkeley) (Accepted for publication, May 29, 1934) 30 years ago, Sir James Walker (1, 2) published two papers entitled, "The theory of amphoteric electrolytes," in which he discussed the conductivities of amphoteric electrolytes and their deviations from the Ostwald dilution law in terms of the mass law. Deriving the expres- sions for the various ion concentrations present in an ampholyte solu- tion, he showed why the attempt to apply the dilution law to such solutions is invalid. He was able to show that the conductivities calculated from the relations he had obtained were in agreement with Winkelblech's (3) data for the aminobermoic acids. With the exception of a single value for asparagine, the figures given by Walker are not concerned with the aliphatic amino acids. At the time there were no trustworthy data for the latter compounds. In 1895, Emile Franke (4) reported values for a-alanine up to a con- centration of 0.0156 ~r, and in 1905, Siegfried (5) published more measurements on alanine, this time from 0.125 to 1.0 ~. Values at isolated concentrations are also given by Bayliss (6). More recently the conductivity of glycine has been measured by Miyamoto and Schmidt (7). In connection with the measurement of the conductivity of asparagine, Walker pointed out the fact that the only criterion for the purity of such a compound is the attainment of constant con- ductivity on repeated purification, and that such a result was only obtained after the asparagine hadbeen recrystallized twenty-four times. -

Asparagine: a Metabolite to Be Targeted in Cancers
H OH metabolites OH Review Asparagine: A Metabolite to Be Targeted in Cancers Jie Jiang 1, Sandeep Batra 2,* and Ji Zhang 1,3,* 1 Herman B Wells Center for Pediatric Research, School of Medicine, Indiana University, Indianapolis, IN 46202, USA; [email protected] 2 Riley Hospital for Children at Indiana University Health; Indianapolis, IN 46202, USA 3 Department of Biochemistry and Molecular Biology, School of Medicine, Indiana University; Indianapolis, IN 46202, USA * Correspondence: [email protected] (S.B.); [email protected] (J.Z.) Abstract: Amino acids play central roles in cancer progression beyond their function as building blocks for protein synthesis. Thus, targeting amino acid acquisition and utilization has been proved to be therapeutically beneficial in various pre-clinical models. In this regard, depletion of circulating asparagine, a nonessential amino acid, by L-asparaginase has been used in treating pediatric acute lymphoblastic leukemia (ALL) for decades. Of interest, unlike most solid tumor cells, ALL cells lack the ability to synthesize their own asparagine de novo effectively. However, only until recently, growing evidence suggests that solid tumor cells strive to acquire adequate amounts of asparagine to support tumor progression. This process is subjected to the regulation at various levels, including oncogenic signal, tumor-niche interaction, intratumor heterogeneity and dietary accessibility. We will review the literature on L-asparaginase-based therapy as well as recent understanding of asparagine metabolism in solid tumor progression, with the hope of shedding light into a broader cancer therapeutic strategy by perturbing its acquisition and utilization. Keywords: asparagine; L-asparaginase; acute lymphoblastic leukemia; asparagine synthetase; stress Citation: Jiang, J.; Batra, S.; Zhang, J. -

24Amino Acids, Peptides, and Proteins
WADEMC24_1153-1199hr.qxp 16-12-2008 14:15 Page 1153 CHAPTER COOϪ a -h eli AMINO ACIDS, x ϩ PEPTIDES, AND NH3 PROTEINS Proteins are the most abundant organic molecules 24-1 in animals, playing important roles in all aspects of cell structure and function. Proteins are biopolymers of Introduction 24A-amino acids, so named because the amino group is bonded to the a carbon atom, next to the carbonyl group. The physical and chemical properties of a protein are determined by its constituent amino acids. The individual amino acid subunits are joined by amide linkages called peptide bonds. Figure 24-1 shows the general structure of an a-amino acid and a protein. α carbon atom O H2N CH C OH α-amino group R side chain an α-amino acid O O O O O H2N CH C OH H2N CH C OH H2N CH C OH H2N CH C OH H2N CH C OH CH3 CH2OH H CH2SH CH(CH3)2 alanine serine glycine cysteine valine several individual amino acids peptide bonds O O O O O NH CH C NH CH C NH CH C NH CH C NH CH C CH3 CH2OH H CH2SH CH(CH3)2 a short section of a protein a FIGURE 24-1 Structure of a general protein and its constituent amino acids. The amino acids are joined by amide linkages called peptide bonds. 1153 WADEMC24_1153-1199hr.qxp 16-12-2008 14:15 Page 1154 1154 CHAPTER 24 Amino Acids, Peptides, and Proteins TABLE 24-1 Examples of Protein Functions Class of Protein Example Function of Example structural proteins collagen, keratin strengthen tendons, skin, hair, nails enzymes DNA polymerase replicates and repairs DNA transport proteins hemoglobin transports O2 to the cells contractile proteins actin, myosin cause contraction of muscles protective proteins antibodies complex with foreign proteins hormones insulin regulates glucose metabolism toxins snake venoms incapacitate prey Proteins have an amazing range of structural and catalytic properties as a result of their varying amino acid composition. -

DNA-Binding Specificity (Protein-DNA Interactions/Bzip Proteins/DNA Sequence Recognition) DIMITRIS TZAMARIAS, WILLIAM T
Proc. Nati. Acad. Sci. USA Vol. 89, pp. 2007-2011, March 1992 Biochemistry Mutations in the bZIP domain of yeast GCN4 that alter DNA-binding specificity (protein-DNA interactions/bZIP proteins/DNA sequence recognition) DIMITRIS TZAMARIAS, WILLIAM T. PU, AND KEVIN STRUHL Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115 Communicated by Stephen C. Harrison, November 18, 1991 (received for review July 30, 1991) ABSTRACT The bZIP class of eukaryotic transcriptional invariant asparagine residue (Asn-235). The two models for regulators utilize a distinct structural motif that consists of a DNA binding by bZIP proteins propose distinct roles for this leucine zipper that mediates dimerization and an adjacent basic asparagine residue. In the scissors-grip model (22), the in- region that directly contacts DNA. Although models of the variant asparagine is proposed to break the a-helix in the protein-DNA complex have been proposed, the basis of DNA- basic region, thus permitting it to bend sharply and wrap binding specificity is essentially unknown. By genetically se- around the DNA. In the induced-fork model (14), the aspar- lecting for derivatives ofyeast GCN4 that activate transcription agine is proposed to directly contact the target sequence. from promoters containing mutant binding sites, we isolate an By genetically selecting for derivatives of GCN4 that can altered-specificity mutant in which the invariant asparagine in activate transcription from promoters containing mutant bind- the basic region of bZIP proteins (Asn-235) has been changed ing sites, we isolate an altered-specificity mutant of yeast to tryptophan. Wild-type GCN4 binds the optimal site (AT- GCN4 in which the invariant asparagine in the basic region of GACTCAT) with much higher affinity than the mutant site bZIP proteins (Asn-235) has been changed to tryptophan. -

Isoleucine, Asparagine, Glutamine, Proline, Leucine, and Glycine Which Bears a Carboxamide Group
OXYTOCIN AND NEUROHYPOPHYSEAL PEPTIDES: SPECTRAL ASSIGNMENT AND CONFORMA TIONAL ANAL YSIS BY 220 M1Hz NUCLEAR MAGNETIC RESONANCE*,t BY LEROY F. JOHNSON, I. L. SCHWARTZ, AND RODERICH WALTERt VARIAN ASSOCIATES, ANALYTICAL INSTRUMENT DIVISION, PALO ALTO, CALIF.; DEPARTMENT OF PHYSIOLOGY, MOUNT SINAI MEDICAL AND GRADUATE SCHOOLS OF THE CITY UNIVERSITY OF NEW YORK; AND MEDICAL RESEARCH CENTER, BROOKHAVEN NATIONAL LABORATORY, UPTON, NEW YORK Communicated by Maurice Goldhaber, May 26, 1969 Abstract. 1\Jagnetic resonance peaks have been assigned to individual protons of the constituent amino acids in the neurohypophyseal hormone, oxytocin, and in related peptides. The assignments were made possible by operation at 220 M\Hz with the use of variable temperature studies, proton homonuclear spin- decoupling, and comparison of spectra of oxytocin analogs. Some of the observed chemical shifts, and NH-CHa coupling constants were studied in relation to the conformation of the hormone. Earlier we investigated the conformation of neurohypophyseal hormones by means of partition chromatography1 and circular dichroism.2 In continuation of these studies we turned to 220 1\IHz proton nuclear magnetic resonance (NM'\1R)- a technique which offers great promise in revealing information about helix-- coil transitions and mobility of side chains as well as intra- and intermolecular interactions in peptides and proteins. In this paper' we wish to report on the analysis of NM'R spectra, in deuterated dimethylsulfoxide, of oxytocin a cyclic peptide hormone composed of eight different amino acids, viz., cystine, tyrosine, isoleucine, asparagine, glutamine, proline, leucine, and glycine which bears a carboxamide group. Materials and Methods.-Spectra were recorded using a Varian Associates HR- 220 spectrometer. -

The Breakdown of Asparagine, Glutamine, and Other Amides by Microorganisms from the Sheep's Rumen
THE BREAKDOWN OF ASPARAGINE, GLUTAMINE, AND OTHER AMIDES BY MICROORGANISMS FROM THE SHEEP'S RUMEN By A. C. 1. W ARNER* [klanuscript received July 17, 1963] Summary Microorganisms from the rumen of sheep rapidly broke down asparagine, glutamine, nicotinamide, and formamide, with the production of ammonia, but only slowly attacked acetamide and propionamide. Microorganisms from different animals, or collected at different times, had different activities. The results suggested that a separate enzyme or enzymes were involved for each substrate, including perhaps a D- as well as an L-asparaginase. The amide groups of casein were also broken down, but it is uncertain to what extent prior hydrolysis had taken place. -while the a·ctivities could not be correlated with any morphologically recognizable group of microorganisms, it appeared that asparaginase was mainly associated with the bacteria, glutaminase to a large extent with the protozoa. The aspartic and glutamic acids formed by dearnidation of asparagine and glutamine were further deaminatod. The optimum pH for aspaJ:'aginase and glutaminase was between 7 and 8, but considera,ble activity remained even at pH 5. Extracts from the microorganisms were made. The asparaginase activity in t.hese was inhibited by mercuric ions and to a lesser extent by ammonium and by cyanide ions. No inhibition or activation was found with phthalein dyes, aspartic acid, phosphate, sulphate, or chloride ions, or toluene. The apparent Km of the asparaginase was less than 10-4. 1\1. 1. INTRODUCTION The natural diet of ruminants contains a considerable variety of nonMprotein nitrogenous material (Chalmers and Synge 1954), and this includes the amino acid amides asparagine and glutamine.