DNA-Binding Specificity (Protein-DNA Interactions/Bzip Proteins/DNA Sequence Recognition) DIMITRIS TZAMARIAS, WILLIAM T
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Amino Acid Recognition by Aminoacyl-Trna Synthetases
www.nature.com/scientificreports OPEN The structural basis of the genetic code: amino acid recognition by aminoacyl‑tRNA synthetases Florian Kaiser1,2,4*, Sarah Krautwurst3,4, Sebastian Salentin1, V. Joachim Haupt1,2, Christoph Leberecht3, Sebastian Bittrich3, Dirk Labudde3 & Michael Schroeder1 Storage and directed transfer of information is the key requirement for the development of life. Yet any information stored on our genes is useless without its correct interpretation. The genetic code defnes the rule set to decode this information. Aminoacyl-tRNA synthetases are at the heart of this process. We extensively characterize how these enzymes distinguish all natural amino acids based on the computational analysis of crystallographic structure data. The results of this meta-analysis show that the correct read-out of genetic information is a delicate interplay between the composition of the binding site, non-covalent interactions, error correction mechanisms, and steric efects. One of the most profound open questions in biology is how the genetic code was established. While proteins are encoded by nucleic acid blueprints, decoding this information in turn requires proteins. Te emergence of this self-referencing system poses a chicken-or-egg dilemma and its origin is still heavily debated 1,2. Aminoacyl-tRNA synthetases (aaRSs) implement the correct assignment of amino acids to their codons and are thus inherently connected to the emergence of genetic coding. Tese enzymes link tRNA molecules with their amino acid cargo and are consequently vital for protein biosynthesis. Beside the correct recognition of tRNA features3, highly specifc non-covalent interactions in the binding sites of aaRSs are required to correctly detect the designated amino acid4–7 and to prevent errors in biosynthesis5,8. -

Electrochemical Studies of Dl-Leucine, L-Proline and L
ELECTROCHEMICAL STUDIES OF DL -LEUCINE, 60 L-PROLINE AND L-TRYPTOPHAN AND THEIR INTERACTION WITH COPPER AND IRON 30 c b A) M. A. Jabbar, R. J. Mannan, S. Salauddin and B. µ a Rashid 0 Department of Chemistry, University of Dhaka, ( Current Dhaka-1000, Bangladesh -30 Introduction -60 In vitro study of the charge transfer reactions coupled -800 -400 0 400 800 with chemical reactions can give important indication of Potential vs. Ag/AgCl (mV) about actual biological processes occurring in human Fig.1. Comparison of the cyclic voltammogram of system. Understanding of such charge-transfer 5.0mM (a) DL -Leucine, (b) Cu-DL -Leucine ion and mechanism will help to determine the effectiveness of (c) [Fe-DL -Leucine] in 0.1M KCl solution at a Pt- nutrition, metabolism and treatment of various biological button electrode. Scan rates 50 mV/s. disorders. In the previous research, the redox behaviour of 40 various amino acids and biochemically important compounds and their charge transfer reaction and their b interaction of metal ions were studied [1,2]. In the present 20 ) a c research, the redox behavior and the charge transfer µΑ kinetics of DL -Leucine, L-Proline and L-Tryptophan in 0 the presence and absence of copper and iron will be investigated. Current ( -20 Experimental A computerized electrochemistry system developed by -40 -800 -400 0 400 800 Advanced Analytics, Virginia, USA, (Model-2040) Potential vs. Ag/AgCl (mV) consisting of three electrodes micro-cell with a saturated Ag/AgCl reference, a Pt-wire auxiliary and a pretreated Fig.2 . Comparison of the cyclic voltammogram of Pt-button working electrode is employed to investigate 5.0mM (a) L-Proline, (b) Cu-L-Proline and (c) Fe-L- Proline in 0.1M KCl solution at a Pt-button different amino acids and metal-amino acid systems. -

Amino Acid Chemistry
Handout 4 Amino Acid and Protein Chemistry ANSC 619 PHYSIOLOGICAL CHEMISTRY OF LIVESTOCK SPECIES Amino Acid Chemistry I. Chemistry of amino acids A. General amino acid structure + HN3- 1. All amino acids are carboxylic acids, i.e., they have a –COOH group at the #1 carbon. 2. All amino acids contain an amino group at the #2 carbon (may amino acids have a second amino group). 3. All amino acids are zwitterions – they contain both positive and negative charges at physiological pH. II. Essential and nonessential amino acids A. Nonessential amino acids: can make the carbon skeleton 1. From glycolysis. 2. From the TCA cycle. B. Nonessential if it can be made from an essential amino acid. 1. Amino acid "sparing". 2. May still be essential under some conditions. C. Essential amino acids 1. Branched chain amino acids (isoleucine, leucine and valine) 2. Lysine 3. Methionine 4. Phenyalanine 5. Threonine 6. Tryptophan 1 Handout 4 Amino Acid and Protein Chemistry D. Essential during rapid growth or for optimal health 1. Arginine 2. Histidine E. Nonessential amino acids 1. Alanine (from pyruvate) 2. Aspartate, asparagine (from oxaloacetate) 3. Cysteine (from serine and methionine) 4. Glutamate, glutamine (from α-ketoglutarate) 5. Glycine (from serine) 6. Proline (from glutamate) 7. Serine (from 3-phosphoglycerate) 8. Tyrosine (from phenylalanine) E. Nonessential and not required for protein synthesis 1. Hydroxyproline (made postranslationally from proline) 2. Hydroxylysine (made postranslationally from lysine) III. Acidic, basic, polar, and hydrophobic amino acids A. Acidic amino acids: amino acids that can donate a hydrogen ion (proton) and thereby decrease pH in an aqueous solution 1. -

Enhancing Effect of Glycine and Tryptophan Mixture on Estimated
Functional Foods in Health and Disease 2021; 11(1): 24-33 www.ffhdj.com Page 24 of 33 Research Article Open Access Enhancing effect of glycine and tryptophan mixture on estimated glomerular filtration rate in healthy participants: A randomized, double-blind, placebo-controlled parallel study Shunji Oshima*, Sachie Shiiya, Yasunori Nakamura Core Technology Laboratories, Asahi Quality and Innovations, Ltd., Ibaraki, Japan *Corresponding author: Shunji Oshima, PhD, Core Technology Laboratories, Asahi Quality & Innovations, Ltd., 1-21, Midori 1-chome, Moriya-shi, Ibaraki, 302-0106, Japan. Submission Date: January 7th, 2021; Acceptance Date: January 22nd, 2021; Publication Date: January 29th, 2021 Please cite this article as: Oshima S., Shiiya S., Nakamura Y. Enhancing effect of glycine and tryptophan mixture on estimated glomerular filtration rate in healthy participants: A randomized, double-blind, placebo-controlled parallel study. Functional Foods in Health and Disease 2021. 11(1): 24-33. DOI: https://www.doi.org/10.31989/ffhd.v11i1.774 ABSTRACT Background: The mixture of glycine and tryptophan exhibited serum uric acid-lowering effects in our previous clinical trial. Objective: Using a randomized, double-blind, placebo- controlled, and parallel study design, this current study aimed to examine whether this mixture enhanced the estimated glomerular filtration rate (eGFR) as an indicator of renal function in healthy individuals. Methods: Healthy Japanese adult males and females ingested a powder mixture containing 3.0 g of glycine and 0.2 g of tryptophan or a placebo powder once daily at bedtime for 8 weeks. Results: After 8 weeks of continual ingestion, the combined glycine and tryptophan supplementation significantly enhanced eGFR. -

Of Net Glutamine Synthesis
Biochem. J. (1991) 277, 697-703 (Printed in Great Britain) 697 Hyperammonaemia does not impair brain function in the absence of net glutamine synthesis Richard A. HAWKINS* and J. JESSY Department of Physiology and Biophysics, The Chicago Medical School, North Chicago, IL 60064, U.S.A. 1. It has been established that chronic hyperammonaemia, whether caused by portacaval shunting or other means, leads to a variety of metabolic changes, including a depression in the cerebral metabolic rate of glucose (CMRGIC), increased permeability of the blood-brain barrier to neutral amino acids, and an increase in the brain content of aromatic amino acids. The preceding paper [Jessy, DeJoseph & Hawkins (1991) Biochem. J. 277, 693-696] showed that the depression in CMRGlC caused by hyperammonaemia correlated more closely with glutamine, a metabolite of ammonia, than with ammonia itself. This suggested that ammonia (NH3 and NH41) was without effect. The present experiments address the question whether ammonia, in the absence of net glutamine synthesis, induces any of the metabolic symptoms of cerebral dysfunction associated with hyperammonaemia. 2. Small doses of methionine sulphoximine, an inhibitor of glutamine synthetase, were used to raise the plasma ammonia levels of normal rats without increasing the brain glutamine content. These hyperammonaemic rats, with plasma and brain ammonia levels equivalent to those known to depress brain function, behaved normally over 48 h. There was no depression of cerebral energy metabolism (i.e. the rate of glucose consumption). Contents of key intermediary metabolites and high-energy phosphates were normal. Neutral amino acid transport (tryptophan and leucine) and the brain contents of aromatic amino acids were unchanged. -

The EWS/ATF1 Fusion Protein Contains a Dispersed Activation Domain That Functions Directly
Oncogene (1998) 16, 1625 ± 1631 1998 Stockton Press All rights reserved 0950 ± 9232/98 $12.00 The EWS/ATF1 fusion protein contains a dispersed activation domain that functions directly Shu Pan, Koh Yee Ming, Theresa A Dunn, Kim KC Li and Kevin AW Lee Department of Biology, Hong Kong University of Science & Technology, Clear Water Bay, Kowloon, Hong Kong, P.R.C. Naturally occurring chromosomal fusion of the Ewings 1994). For all of the above malignancies, the EWS Sarcoma Oncogene (EWS) to distinct cellular transcrip- fusion proteins function as potent transcriptional tion factors, produces aberrant transcriptional activators activators (May et al., 1993b; Ohno et al., 1993; that function as dominant oncogenes. In Malignant Bailly et al., 1994; Brown et al., 1995; Lessnick et al., Melanoma of Soft Parts the N-terminal region of 1995; Fujimura et al., 1996) in a manner that is EWS is fused to C-terminal region of the cAMP- dependent on the EWS N-terminal region, hereafter inducible transcription factor ATF1. The EWS/ATF1 referred to as the EWS Activation Domain (EAD). It is fusion protein binds to ATF sites present in cAMP- envisioned that distinct tumors arise via de-regulation responsive promoters via the ATF1 bZIP domain and of dierent genes, depending on the fusion partner for activates transcription constitutively in a manner that is EWS. In cases where it has been examined, agents that dependent on an activation domain (EAD) present in antagonise EWS-fusion proteins also inhibit cellular EWS. To further de®ne the requirements for trans- proliferation (Ouchida et al., 1995; Kovar et al., 1996; activation we have performed mutational analysis of Yi et al., 1997; Tanaka et al., 1997), indicating that EWS/ATF1 in mammalian cells and report several new EWS fusions can play a role in both tumor formation ®ndings. -

A Placebo Controlled Investigation of the Effects of Tryptophan Or Placebo on Subjective and Objective Measures of Fatigue
European Journal of Clinical Nutrition (1998) 52, 425±431 ß 1998 Stockton Press. All rights reserved 0954±3007/98 $12.00 http://www.stockton-press.co.uk/ejcn A placebo controlled investigation of the effects of tryptophan or placebo on subjective and objective measures of fatigue A Cunliffe, OA Obeid and J Powell-Tuck Department of Human Nutrition, St Bartholomew's and Royal London School of Medicine and Dentistry, Queen Mary and West®eld College, London E1 2AD Objective: To examine the effect of L-tryptophan administration on subjective and objective measures of fatigue in healthy volunteers. Subjects: Six healthy volunteers (4M:2F) were recruited from staff and students at the College. Setting: Department of Human Nutrition, St. Bartholomews and the Royal London School of Medicine and Dentistry. Design: Subjects were tested for central and peripheral fatigue using a visual analogue scale, ¯icker fusion frequency, grip strength, reaction time and wrist ergometry. In addition, plasma free tryptophan concentrations and Trp:LNAA ratio were determined. Measurements were made before, and at 1, 2, 3 and 4 h after drinking one of two test drinks. The drinks were of either caffeine free diet Coca-Cola (placebo) or caffeine free diet Coca- Cola plus L-tryptophan (30 mg/kg: active drink). Each of the six subjects was tested after placebo and active drink with a one week washout period between test days. Results: Subjective fatigue was signi®cantly increased following tryptophan compared to placebo (P < 0.002), and objective measures of central fatigue were signi®cantly increased by tryptophan compared to placebo (¯icker fusion frequency: P < 0.001; reaction time P < 0.001). -

Nucleotide Base Coding and Am1ino Acid Replacemients in Proteins* by Emil L
VOL. 48, 1962 BIOCHEMISTRY: E. L. SAIITH 677 18 Britten, R. J., and R. B. Roberts, Science, 131, 32 (1960). '9 Crestfield, A. M., K. C. Smith, and F. WV. Allen, J. Biol. Chem., 216, 185 (1955). 20 Gamow, G., Nature, 173, 318 (1954). 21 Brenner, S., these PROCEEDINGS, 43, 687 (1957). 22 Nirenberg, M. WV., J. H. Matthaei, and 0. WV. Jones, unpublished data. 23 Crick, F. H. C., L. Barnett, S. Brenner, and R. J. Watts-Tobin, Nature, 192, 1227 (1961). 24 Levene, P. A., and R. S. Tipson, J. Biol. Ch-nn., 111, 313 (1935). 25 Gierer, A., and K. W. Mundry, Nature, 182, 1437 (1958). 2' Tsugita, A., and H. Fraenkel-Conrat, J. Mllot. Biol., in press. 27 Tsugita, A., and H. Fraenkel-Conrat, personal communication. 28 Wittmann, H. G., Naturwissenschaften, 48, 729 (1961). 29 Freese, E., in Structure and Function of Genetic Elements, Brookhaven Symposia in Biology, no. 12 (1959), p. 63. NUCLEOTIDE BASE CODING AND AM1INO ACID REPLACEMIENTS IN PROTEINS* BY EMIL L. SMITHt LABORATORY FOR STUDY OF HEREDITARY AND METABOLIC DISORDERS AND THE DEPARTMENTS OF BIOLOGICAL CHEMISTRY AND MEDICINE, UNIVERSITY OF UTAH COLLEGE OF MEDICINE Communicated by Severo Ochoa, February 14, 1962 The problem of which bases of messenger or template RNA' specify the coding of amino acids in proteins has been largely elucidated by the use of synthetic polyri- bonucleotides.2-7 For these triplet nucleotide compositions (Table 1), it is of in- terest to examine some of the presently known cases of amino acid substitutions in polypeptides or proteins of known structure. -

An Integrated Meta-Analysis of Peripheral Blood Metabolites and Biological Functions in Major Depressive Disorder
Molecular Psychiatry https://doi.org/10.1038/s41380-020-0645-4 ARTICLE An integrated meta-analysis of peripheral blood metabolites and biological functions in major depressive disorder 1,2,3 1,2,3 1,2,3 1,3 1,3 4,5 1,3 1,3 Juncai Pu ● Yiyun Liu ● Hanping Zhang ● Lu Tian ● Siwen Gui ● Yue Yu ● Xiang Chen ● Yue Chen ● 1,2,3 1,3 1,3 1,3 1,3 1,2,3 Lining Yang ● Yanqin Ran ● Xiaogang Zhong ● Shaohua Xu ● Xuemian Song ● Lanxiang Liu ● 1,2,3 1,3 1,2,3 Peng Zheng ● Haiyang Wang ● Peng Xie Received: 3 June 2019 / Revised: 24 December 2019 / Accepted: 10 January 2020 © The Author(s) 2020. This article is published with open access Abstract Major depressive disorder (MDD) is a serious mental illness, characterized by high morbidity, which has increased in recent decades. However, the molecular mechanisms underlying MDD remain unclear. Previous studies have identified altered metabolic profiles in peripheral tissues associated with MDD. Using curated metabolic characterization data from a large sample of MDD patients, we meta-analyzed the results of metabolites in peripheral blood. Pathway and network analyses were then performed to elucidate the biological themes within these altered metabolites. We identified 23 differentially 1234567890();,: 1234567890();,: expressed metabolites between MDD patients and controls from 46 studies. MDD patients were characterized by higher levels of asymmetric dimethylarginine, tyramine, 2-hydroxybutyric acid, phosphatidylcholine (32:1), and taurochenode- soxycholic acid and lower levels of L-acetylcarnitine, creatinine, L-asparagine, L-glutamine, linoleic acid, pyruvic acid, palmitoleic acid, L-serine, oleic acid, myo-inositol, dodecanoic acid, L-methionine, hypoxanthine, palmitic acid, L-tryptophan, kynurenic acid, taurine, and 25-hydroxyvitamin D compared with controls. -

Impact of Tryptophan and Glutamine on the Tissue Culture of Upland Rice
Impact of tryptophan and glutamine on the tissue culture of upland rice E. Shahsavari School of Biological Sciences, Flinders University, Adelaide, Australia ABSTRACT In order to evaluate the effect of tryptophan and glutamine on the tissue culture of upland rice cultivars, serial ex- periments were conducted using four cultivars: Kusan, Lamsan, Selasi and Siam. Mature seeds from these cultivars were subjected to 4 levels of tryptophan and glutamine on the MSB5 (MS macro elements, B5 micro elements and B5 vitamins) medium. Callus induction results showed a positive effect of tryptophan on all cultivars except Selasi. The optimal tryptophan concentration for callus induction in cultivars Kusan and Siam was 100 µmol, while in Lamsan the optimum was 200 µmol. With the exception of the Lamsan cultivar, incorporation of glutamine gener- ally did not result in the enhancement of callus induction response that incorporation of tryptophan did. Plantlet regeneration frequency was significantly increased when an appropriate level of tryptophan was added to culture media, the optimum being 100 µmol or Kusan, Selasi and Siam, compared to an optimum of 200 µmol for Lamsan. Glutamine did not affect regeneration frequency in any of the cultivars under the conditions tested. In summary, the results showed that tryptophan is a useful additive for upland rice tissue culture. Keywords: callus induction frequency; plantlet regeneration frequency; embryogenic calli Around the globe, rice is a very important crop The previous investigations showed the positive as a staple food and a model plant for genomic effect of tryptophan on the tissue culture system study (Tyagi and Mohanty 2000, Bajaj and Mohanty of rice (Siriwardana and Nabors 1983, Chowdhry 2005). -
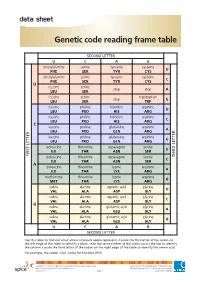
Genetic Code Reading Frame Table
data sheet Genetic code reading frame table SECOND LETTER U C A G phenylalanine serine tyrosine cysteine U PHE SER TYR CYS phenylalanine serine tyrosine cysteine C PHE SER TYR CYS U leucine serine stop stop A LEU SER leucine serine tryptophan stop G LEU SER TRP leucine proline histidine arginine U LEU PRO HIS ARG leucine proline histidine arginine C LEU PRO HIS ARG C leucine proline glutamine arginine A LEU PRO GLN ARG leucine proline glutamine arginine G LEU PRO GLN ARG isoleucine threonine asparagine serine U ILE THR ASN SER FIRST LETTER isoleucine threonine asparagine serine LETTERTHIRD C ILE THR ASN SER A isoleucine threonine lysine arginine A ILE THR LYS ARG methionine threonine lysine arginine G MET THR LYS ARG valine alanine aspartic acid glycine U VAL ALA ASP GLY valine alanine aspartic acid glycine C VAL ALA ASP GLY G valine alanine glutamic acid glycine A VAL ALA GLU GLY valine alanine glutamic acid glycine G VAL ALA GLU GLY U C A G SECOND LETTER Use this table to find out what amino acid each codon represents. Locate the first letter of the codon on the left edge of the table to identify a block, then the second letter of the codon across the top to identify the column. Locate the third letter of the codon on the right edge of the table to identify the amino acid. For example, the codon ‘CAU’ codes for histidine (HIS). ast0812 | Proteins 3: Genetic code reading frame table (data sheet) developed for the Department of Education WA © The University of Western Australia 2012 for conditions of use see spice.wa.edu.au/usage version 1.0 page 1 Licensed for NEALS. -

Analysis of Amino Acids by HPLC
Analysis of Amino Acids by HPLC Rita Steed Agilent Technologies, Inc. 800-227-9770 opt 3/opt3/opt 2 Amino Acid Analysis - Agilent Restricted Page 1 June 24, 2010 Outline • Amino Acids – Structure, Chemistry • Separation Considerations • Challenges • Instrumentation • Derivatization – OPA, FMOC • Overview of Separations • Examples Amino Acid Analysis - Agilent Restricted Page 2 June 24, 2010 Amino Acids – Structure, Chemistry CH3 Alanine (()Ala) Glutamic Acid (()Glu) Amino Acid Analysis - Agilent Restricted Page 3 June 24, 2010 Amino Acids – Zwitterionic Amino Acid Analysis - Agilent Restricted Page 4 June 24, 2010 Separation Considerations • Zwitterions - poor solubility near iso- …electric point • Most have poor UV absorbance • Derivatization – OPA, FMOC •Reduce polarity – increases retention in reversed-phase chromatoggpyraphy •Improve sensitivity – UV, Fluorescence • Detector; DAD, FLD, MS, ELSD Amino Acid Analysis - Agilent Restricted Page 5 June 24, 2010 Ortho Phthalaldehyde (OPA) and Fluorenylmethoxy chloroformate (FMOC) Reactions with Amines OPA O SR’ R’SH H NR +RNH2 H Room Temperature O Fluorescence: Ex 340nm, Em 450nm Non-fluorescent DAD: 338 , 10nm; Ref . 390 , 20nm Does not absorb at 338nm FMOC RR’NH - HCl + or Room Temperature RNH2 NRR’ or NHR Fluorescence: Ex 260nm, Em 325nm Fluorescent DAD: 262, 16nm; Ref. 324,8nm Absorbs at 262nm and Fluorescences at 324nm Group/Presentation Title Agilent Restricted Month ##, 200X Names and Order of Elution for OPA and FMOC Derivatives of Amino Acids Peak # AA Name AA Abbreviation Derivative Type Peak # AA Name AA Abbreviation Derivative Type Group/Presentation Title Agilent Restricted Month ##, 200X Agilent AAA Methods - They’ve Evolved • Automated Amino Acid Analysis – AminoQuant I & II (1987) •1090 •1100, Pub.