Independent Highly Sensitive Characterization of Asparagine
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Deamidation of Human Proteins
Deamidation of human proteins N. E. Robinson*† and A. B. Robinson‡ *Division of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, CA 91125; and ‡Oregon Institute of Science and Medicine, Cave Junction, OR 97523 Communicated by Frederick Seitz, The Rockefeller University, New York, NY, August 31, 2001 (received for review May 8, 2001) Deamidation of asparaginyl and glutaminyl residues causes time- 3D structure is known (23). This method is more than 95% dependent changes in charge and conformation of peptides and reliable in predicting relative deamidation rates of Asn residues proteins. Quantitative and experimentally verified predictive cal- within a single protein and is also useful for the prediction of culations of the deamidation rates of 1,371 asparaginyl residues in absolute deamidation rates. a representative collection of 126 human proteins have been It is, therefore, now possible to compute the expected deami- performed. These rates suggest that deamidation is a biologically dation rate of any protein for which the primary and 3D relevant phenomenon in a remarkably large percentage of human structures are known, except for very long-lived proteins. These proteins. proteins require measurement of the 400 Gln pentapeptide rates. in vivo deamidation ͉ asparaginyl residues Materials and Methods Calculation Method. The Brookhaven Protein Data Bank (PDB) eamidation of asparaginyl (Asn) and glutaminyl (Gln) was searched to select 126 human proteins of general biochem- Dresidues to produce aspartyl (Asp) and glutamyl (Glu) ical interest and of known 3D structure without bias toward any residues causes structurally and biologically important alter- known data about their deamidation, except for 13 proteins (as ations in peptide and protein structures. -

Deamidation, Acylation and Proteolysis of a Model Peptide in PLGA Films ⁎ M.L
Journal of Controlled Release 112 (2006) 111–119 www.elsevier.com/locate/jconrel Deamidation, acylation and proteolysis of a model peptide in PLGA films ⁎ M.L. Houchin a, K. Heppert b, E.M. Topp a, a Department of Pharmaceutical Chemistry, The University of Kansas, 2095 Constant Ave., Lawrence, KS 66047, United States b Higuchi Biosciences Centers, The University of Kansas, Lawrence, KS, United States Received 8 December 2005; accepted 30 January 2006 Available online 9 March 2006 Abstract The relative rates of deamidation, acylation and proteolysis (i.e. amide bond cleavage) were determined for a model peptide (VYPNGA) in poly (DL-lactide-co-glycolide) films. Films were stored at 70°C and either 95%, 75%, 60%, 45%, 28%, or ∼0% relative humidity and at 37°C and 95% relative humidity. Peptide degradation products were identified by ESI+MS/MS and quantitated by LC/MS/MS. Extensive overlap of degradation mechanisms occurred, producing a complex mixture of products. Acylation was the dominant peptide degradation reaction (10–20% of total peptide) at early stages of PLGA hydrolysis and at intermediate relative humidity (60–45% RH). Deamidation and proteolysis were dominant (25–50% and 20–40% of total peptide, respectively) at later stages and at high relative humidity (95–75% RH). Understanding the relative rates of each peptide degradation reaction will allow for improved design of PLGA formulations that preserve the stability of peptide and protein drugs. © 2006 Elsevier B.V. All rights reserved. Keywords: PLGA; Deamidation; Acylation; Proteolysis; Peptide stability 1. Introduction bonds produces lactic and glycolic acid, which are easily metabolized. -

Amino Acid Chemistry
Handout 4 Amino Acid and Protein Chemistry ANSC 619 PHYSIOLOGICAL CHEMISTRY OF LIVESTOCK SPECIES Amino Acid Chemistry I. Chemistry of amino acids A. General amino acid structure + HN3- 1. All amino acids are carboxylic acids, i.e., they have a –COOH group at the #1 carbon. 2. All amino acids contain an amino group at the #2 carbon (may amino acids have a second amino group). 3. All amino acids are zwitterions – they contain both positive and negative charges at physiological pH. II. Essential and nonessential amino acids A. Nonessential amino acids: can make the carbon skeleton 1. From glycolysis. 2. From the TCA cycle. B. Nonessential if it can be made from an essential amino acid. 1. Amino acid "sparing". 2. May still be essential under some conditions. C. Essential amino acids 1. Branched chain amino acids (isoleucine, leucine and valine) 2. Lysine 3. Methionine 4. Phenyalanine 5. Threonine 6. Tryptophan 1 Handout 4 Amino Acid and Protein Chemistry D. Essential during rapid growth or for optimal health 1. Arginine 2. Histidine E. Nonessential amino acids 1. Alanine (from pyruvate) 2. Aspartate, asparagine (from oxaloacetate) 3. Cysteine (from serine and methionine) 4. Glutamate, glutamine (from α-ketoglutarate) 5. Glycine (from serine) 6. Proline (from glutamate) 7. Serine (from 3-phosphoglycerate) 8. Tyrosine (from phenylalanine) E. Nonessential and not required for protein synthesis 1. Hydroxyproline (made postranslationally from proline) 2. Hydroxylysine (made postranslationally from lysine) III. Acidic, basic, polar, and hydrophobic amino acids A. Acidic amino acids: amino acids that can donate a hydrogen ion (proton) and thereby decrease pH in an aqueous solution 1. -

Nobel Lecture by Roger Y. Tsien
CONSTRUCTING AND EXPLOITING THE FLUORESCENT PROTEIN PAINTBOX Nobel Lecture, December 8, 2008 by Roger Y. Tsien Howard Hughes Medical Institute, University of California San Diego, 9500 Gilman Drive, La Jolla, CA 92093-0647, USA. MOTIVATION My first exposure to visibly fluorescent proteins (FPs) was near the end of my time as a faculty member at the University of California, Berkeley. Prof. Alexander Glazer, a friend and colleague there, was the world’s expert on phycobiliproteins, the brilliantly colored and intensely fluorescent proteins that serve as light-harvesting antennae for the photosynthetic apparatus of blue-green algae or cyanobacteria. One day, probably around 1987–88, Glazer told me that his lab had cloned the gene for one of the phycobilipro- teins. Furthermore, he said, the apoprotein produced from this gene became fluorescent when mixed with its chromophore, a small molecule cofactor that could be extracted from dried cyanobacteria under conditions that cleaved its bond to the phycobiliprotein. I remember becoming very excited about the prospect that an arbitrary protein could be fluorescently tagged in situ by genetically fusing it to the phycobiliprotein, then administering the chromophore, which I hoped would be able to cross membranes and get inside cells. Unfortunately, Glazer’s lab then found out that the spontane- ous reaction between the apoprotein and the chromophore produced the “wrong” product, whose fluorescence was red-shifted and five-fold lower than that of the native phycobiliprotein1–3. An enzyme from the cyanobacteria was required to insert the chromophore correctly into the apoprotein. This en- zyme was a heterodimer of two gene products, so at least three cyanobacterial genes would have to be introduced into any other organism, not counting any gene products needed to synthesize the chromophore4. -

BOSTON UNIVERSITY SCHOOL of MEDICINE Dissertation
BOSTON UNIVERSITY SCHOOL OF MEDICINE Dissertation DEAMIDATION AND RELATED PROBLEMS IN STRUCTURAL ANALYSIS OF PEPTIDES AND PROTEINS by NADEZDA P. SARGAEVA B.S., St. Petersburg State Polytechnical University, 2002 M.S., St. Petersburg State Polytechnical University, 2004 Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy 2012 Approved by First Reader Peter B. O’Connor, Ph.D. Associate Professor of Biochemistry Second Reader Cheng Lin, Ph.D. Assistant Professor of Biochemistry Dedications I dedicate this thesis to my Family iii Acknowledgments I would like to express my deepest gratitude to my advisor, Prof. Peter B. O’Connor. We met in St. Petersburg, Russia in 2004 and you believed in my abilities and you gave me an opportunity to be a part of such a great mass spectrometry program. I also want to thank you for your support and your guidance throughout my Ph.D. studies. Thank you for pushing me to achieve my goals of learning and becoming a better scientist, and for the opportunity you gave me to build up my professional and scientific network. Your help, your advice, and your encouragement in building up my professional career are very much appreciated. Prof. Cheng Lin, thank you for being my second reader and mentor. Working with you has always been a pleasure – I highly value your well thought out ideas, your constant use of the scientific principle, and your willingness to follow up regarding the data, the future directions, etc. I truly appreciate your attention to details, your regular availability and willingness to help me hands on. -

Nucleotide Base Coding and Am1ino Acid Replacemients in Proteins* by Emil L
VOL. 48, 1962 BIOCHEMISTRY: E. L. SAIITH 677 18 Britten, R. J., and R. B. Roberts, Science, 131, 32 (1960). '9 Crestfield, A. M., K. C. Smith, and F. WV. Allen, J. Biol. Chem., 216, 185 (1955). 20 Gamow, G., Nature, 173, 318 (1954). 21 Brenner, S., these PROCEEDINGS, 43, 687 (1957). 22 Nirenberg, M. WV., J. H. Matthaei, and 0. WV. Jones, unpublished data. 23 Crick, F. H. C., L. Barnett, S. Brenner, and R. J. Watts-Tobin, Nature, 192, 1227 (1961). 24 Levene, P. A., and R. S. Tipson, J. Biol. Ch-nn., 111, 313 (1935). 25 Gierer, A., and K. W. Mundry, Nature, 182, 1437 (1958). 2' Tsugita, A., and H. Fraenkel-Conrat, J. Mllot. Biol., in press. 27 Tsugita, A., and H. Fraenkel-Conrat, personal communication. 28 Wittmann, H. G., Naturwissenschaften, 48, 729 (1961). 29 Freese, E., in Structure and Function of Genetic Elements, Brookhaven Symposia in Biology, no. 12 (1959), p. 63. NUCLEOTIDE BASE CODING AND AM1INO ACID REPLACEMIENTS IN PROTEINS* BY EMIL L. SMITHt LABORATORY FOR STUDY OF HEREDITARY AND METABOLIC DISORDERS AND THE DEPARTMENTS OF BIOLOGICAL CHEMISTRY AND MEDICINE, UNIVERSITY OF UTAH COLLEGE OF MEDICINE Communicated by Severo Ochoa, February 14, 1962 The problem of which bases of messenger or template RNA' specify the coding of amino acids in proteins has been largely elucidated by the use of synthetic polyri- bonucleotides.2-7 For these triplet nucleotide compositions (Table 1), it is of in- terest to examine some of the presently known cases of amino acid substitutions in polypeptides or proteins of known structure. -

An Integrated Meta-Analysis of Peripheral Blood Metabolites and Biological Functions in Major Depressive Disorder
Molecular Psychiatry https://doi.org/10.1038/s41380-020-0645-4 ARTICLE An integrated meta-analysis of peripheral blood metabolites and biological functions in major depressive disorder 1,2,3 1,2,3 1,2,3 1,3 1,3 4,5 1,3 1,3 Juncai Pu ● Yiyun Liu ● Hanping Zhang ● Lu Tian ● Siwen Gui ● Yue Yu ● Xiang Chen ● Yue Chen ● 1,2,3 1,3 1,3 1,3 1,3 1,2,3 Lining Yang ● Yanqin Ran ● Xiaogang Zhong ● Shaohua Xu ● Xuemian Song ● Lanxiang Liu ● 1,2,3 1,3 1,2,3 Peng Zheng ● Haiyang Wang ● Peng Xie Received: 3 June 2019 / Revised: 24 December 2019 / Accepted: 10 January 2020 © The Author(s) 2020. This article is published with open access Abstract Major depressive disorder (MDD) is a serious mental illness, characterized by high morbidity, which has increased in recent decades. However, the molecular mechanisms underlying MDD remain unclear. Previous studies have identified altered metabolic profiles in peripheral tissues associated with MDD. Using curated metabolic characterization data from a large sample of MDD patients, we meta-analyzed the results of metabolites in peripheral blood. Pathway and network analyses were then performed to elucidate the biological themes within these altered metabolites. We identified 23 differentially 1234567890();,: 1234567890();,: expressed metabolites between MDD patients and controls from 46 studies. MDD patients were characterized by higher levels of asymmetric dimethylarginine, tyramine, 2-hydroxybutyric acid, phosphatidylcholine (32:1), and taurochenode- soxycholic acid and lower levels of L-acetylcarnitine, creatinine, L-asparagine, L-glutamine, linoleic acid, pyruvic acid, palmitoleic acid, L-serine, oleic acid, myo-inositol, dodecanoic acid, L-methionine, hypoxanthine, palmitic acid, L-tryptophan, kynurenic acid, taurine, and 25-hydroxyvitamin D compared with controls. -

Deamidation in Moxetumomab Pasudotox Leading to Conformational Change And
bioRxiv preprint doi: https://doi.org/10.1101/761635; this version posted September 8, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Deamidation in Moxetumomab Pasudotox Leading to Conformational Change and Immunotoxin Activity Loss X. Lu, S. Lin, N. De Mel, A. Parupudi, M. Pandey, J. Delmar, X. Wang, and J. Wang Running Title: Deamidation impacts immunotoxin activity 1 bioRxiv preprint doi: https://doi.org/10.1101/761635; this version posted September 8, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. ABSTRACT Asparagine deamidation is a common posttranslational modification in which asparagine is converted to aspartic acid or isoaspartic acid. By introducing a negative charge, deamidation could potentially impact the binding interface and biological activities of protein therapeutics. We identified a deamidation variant in moxetumomab pasudotox, an immunotoxin Fv fusion protein drug derived from a 38-kilodalton (kDa) truncated Pseudomonas exotoxin A (PE38) for the treatment of hairy-cell leukemia. Although the deamidation site, Asn-358, was outside of the binding interface, the modification had a significant impact on the biological activity of moxetumomab pasudotox. Surprisingly, the variant eluted earlier than its unmodified form on anion exchange chromatography, which suggests a higher positive charge. -

Modeling the Deamidation of Asparagine Residues Via Succinimide Intermediates
J Mol Model (2001) 7:147–160 DOI 10.1007/s008940100025 ORIGINAL PAPER F. Aylin (Sungur) Konuklar · Viktorya Aviyente Taner Zafer Sen · Ivet Bahar Modeling the deamidation of asparagine residues via succinimide intermediates Received: 29 June 2000 / Accepted: 22 March 2001 / Published online: 24 May 2001 © Springer-Verlag 2001 Abstract Density functional theory (B3LYP/6-31G*) Asp) in a ratio of 1:3. [6] In addition, small amounts of has been used to study the cyclization, deamidation and D-aspartic acid and D-isoaspartic acid were observed, in- hydrolysis reactions of a model peptide. Single point en- dicating slow racemization of the cyclic imide. ergy calculations with the polarized continuum model Spontaneous deamidation through the cyclization drastically lower the activation energy for cyclization in mechanism has been implicated in the inactivation of a a basic medium. Confirmation of the experimental re- number of enzymes. [7] The deamidation reaction has a sults that cyclization is slower than deamidation in acidic much shorter half-life than that for peptide bond hydroly- media and the opposite is true in basic media has enabled sis, varying from days to weeks depending on the amino us to propose mechanisms for both processes. acid sequence. [3] Antibodies that catalyze the deamida- tion and β-aspartyl shift of modified asparaginyl–glycyl Keywords Deamidation · Succinimide · Density dipeptides have been reported. [8] Two classes of anti- functional theory · Hydrolysis of peptides bodies that control theisoaspartate to aspartate product ra- tio were generated. One class catalyzes only the hydroly- sis of succinimide and the other catalyzes both the rate- Introduction limiting deamidation and the subsequent succinimide hy- drolysis. -

Can the Fact That Myelin Proteins Are Old and Break Down Explain the Origin of Multiple Sclerosis in Some People?
Journal of Clinical Medicine Review Can the Fact That Myelin Proteins Are Old and Break down Explain the Origin of Multiple Sclerosis in Some People? Roger J. W. Truscott * and Michael G. Friedrich Illawarra Health and Medical Research Institute, University of Wollongong, Wollongong, NSW 2522, Australia; [email protected] * Correspondence: [email protected]; Tel.: +61-2-4298-3503; Fax: +61-2-4221-8130 Received: 11 July 2018; Accepted: 12 September 2018; Published: 14 September 2018 Abstract: Recent discoveries may change the way that multiple sclerosis (MS) is viewed, particularly with regard to the reasons for the untoward immune response. The fact that myelin proteins are long-lived, and that by the time we are adults, they are extensively degraded, alters our perspective on the reasons for the onset of autoimmunity and the origin of MS. For example, myelin basic protein (MBP) from every human brain past the age of 20 years, is so greatly modified, that it is effectively a different protein from the one that was laid down in childhood. Since only a subset of people with such degraded MBP develop MS, a focus on understanding the mechanism of immune responses to central nervous system (CNS) antigens and cerebral immune tolerance appear to be worthwhile avenues to explore. In accord with this, it will be productive to examine why all people, whose brains contain large quantities of a “foreign antigen”, do not develop MS. Importantly for the potential causation of MS, MBP from MS patients breaks down differently from the MBP in aged controls. If the novel structures formed in these MS-specific regions are particularly antigenic, it could help explain the origin of MS. -
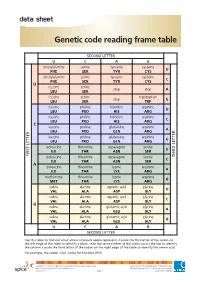
Genetic Code Reading Frame Table
data sheet Genetic code reading frame table SECOND LETTER U C A G phenylalanine serine tyrosine cysteine U PHE SER TYR CYS phenylalanine serine tyrosine cysteine C PHE SER TYR CYS U leucine serine stop stop A LEU SER leucine serine tryptophan stop G LEU SER TRP leucine proline histidine arginine U LEU PRO HIS ARG leucine proline histidine arginine C LEU PRO HIS ARG C leucine proline glutamine arginine A LEU PRO GLN ARG leucine proline glutamine arginine G LEU PRO GLN ARG isoleucine threonine asparagine serine U ILE THR ASN SER FIRST LETTER isoleucine threonine asparagine serine LETTERTHIRD C ILE THR ASN SER A isoleucine threonine lysine arginine A ILE THR LYS ARG methionine threonine lysine arginine G MET THR LYS ARG valine alanine aspartic acid glycine U VAL ALA ASP GLY valine alanine aspartic acid glycine C VAL ALA ASP GLY G valine alanine glutamic acid glycine A VAL ALA GLU GLY valine alanine glutamic acid glycine G VAL ALA GLU GLY U C A G SECOND LETTER Use this table to find out what amino acid each codon represents. Locate the first letter of the codon on the left edge of the table to identify a block, then the second letter of the codon across the top to identify the column. Locate the third letter of the codon on the right edge of the table to identify the amino acid. For example, the codon ‘CAU’ codes for histidine (HIS). ast0812 | Proteins 3: Genetic code reading frame table (data sheet) developed for the Department of Education WA © The University of Western Australia 2012 for conditions of use see spice.wa.edu.au/usage version 1.0 page 1 Licensed for NEALS. -

Analysis of Amino Acids by HPLC
Analysis of Amino Acids by HPLC Rita Steed Agilent Technologies, Inc. 800-227-9770 opt 3/opt3/opt 2 Amino Acid Analysis - Agilent Restricted Page 1 June 24, 2010 Outline • Amino Acids – Structure, Chemistry • Separation Considerations • Challenges • Instrumentation • Derivatization – OPA, FMOC • Overview of Separations • Examples Amino Acid Analysis - Agilent Restricted Page 2 June 24, 2010 Amino Acids – Structure, Chemistry CH3 Alanine (()Ala) Glutamic Acid (()Glu) Amino Acid Analysis - Agilent Restricted Page 3 June 24, 2010 Amino Acids – Zwitterionic Amino Acid Analysis - Agilent Restricted Page 4 June 24, 2010 Separation Considerations • Zwitterions - poor solubility near iso- …electric point • Most have poor UV absorbance • Derivatization – OPA, FMOC •Reduce polarity – increases retention in reversed-phase chromatoggpyraphy •Improve sensitivity – UV, Fluorescence • Detector; DAD, FLD, MS, ELSD Amino Acid Analysis - Agilent Restricted Page 5 June 24, 2010 Ortho Phthalaldehyde (OPA) and Fluorenylmethoxy chloroformate (FMOC) Reactions with Amines OPA O SR’ R’SH H NR +RNH2 H Room Temperature O Fluorescence: Ex 340nm, Em 450nm Non-fluorescent DAD: 338 , 10nm; Ref . 390 , 20nm Does not absorb at 338nm FMOC RR’NH - HCl + or Room Temperature RNH2 NRR’ or NHR Fluorescence: Ex 260nm, Em 325nm Fluorescent DAD: 262, 16nm; Ref. 324,8nm Absorbs at 262nm and Fluorescences at 324nm Group/Presentation Title Agilent Restricted Month ##, 200X Names and Order of Elution for OPA and FMOC Derivatives of Amino Acids Peak # AA Name AA Abbreviation Derivative Type Peak # AA Name AA Abbreviation Derivative Type Group/Presentation Title Agilent Restricted Month ##, 200X Agilent AAA Methods - They’ve Evolved • Automated Amino Acid Analysis – AminoQuant I & II (1987) •1090 •1100, Pub.