Lodothyronine 5'-Deiodinase in Rat Kidney Microsomes Kinetic Behavior at Low Substrate Concentrations Ajit Goswami and Isadore N
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Metabolism of Reverse Triiodothyronine by Isolated Rat Hepatocytes
Metabolism of reverse triiodothyronine by isolated rat hepatocytes. S J Rooda, … , M A van Loon, T J Visser J Clin Invest. 1987;79(6):1740-1748. https://doi.org/10.1172/JCI113014. Research Article Reverse triiodothyronine (rT3) is metabolized predominantly by outer ring deiodination to 3,3'-diiodothyronine (3,3'-T2) in the liver. Metabolism of rT3 and 3,3'-T2 by isolated rat hepatocytes was analyzed by Sephadex LH-20 chromatography, high performance liquid chromatography, and radioimmunoassay, with closely agreeing results. Deiodinase activity was inhibited with propylthiouracil (PTU) and sulfotransferase activity by sulfate depletion or addition of salicylamide or dichloronitrophenol. Normally, little 3,3'-T2 production from rT3 was observed, and 125I- was the main product of both 3, [3'-125I]T2 and [3',5'-125I]rT3. PTU inhibited rT3 metabolism but did not affect 3,3'-T2 clearance as explained by accumulation of 3,3'-T2 sulfate. Inhibition of sulfation did not affect rT3 clearance but 3,3'-T2 metabolism was greatly diminished. The decrease in I- formation from rT3 was compensated by an increased recovery of 3,3'-T2 up to 70% of rT3 metabolized. In conclusion, significant production of 3,3'-T2 from rT3 by rat hepatocytes is only observed if further sulfation is inhibited. Find the latest version: https://jci.me/113014/pdf Metabolism of Reverse Triiodothyronine by Isolated Rat Hepatocytes Sebo Jan Eelkman Rooda, Maria A. C. van Loon, and Thoo J. Vissef Department ofInternal Medicine III and Clinical Endocrinology, Erasmus University Medical School, Rotterdam, The Netherlands Abstract It deiodinates only the outer ring of substrates such as T4 and rT3 (2-4). -

United States Patent (10) Patent N0.: US 7,288,257 B2 Powell (45) Date of Patent: Oct
US007288257B2 (12) United States Patent (10) Patent N0.: US 7,288,257 B2 Powell (45) Date of Patent: Oct. 30, 2007 (54) DIAGNOSIS AND TREATMENT OF HUMAN 5,342,788 A 8/1994 Kunst et a1. .............. .. 436/500 DORMANCY SYNDROME 5,691,456 A 11/1997 AdamcZyk et al. ....... .. 530/405 6,087,090 A 7/2000 Mascarenhas ................ .. 435/4 (76) Inventor: Michael Powell, 150 Catherine Lance, 2003/0007941 A1 1/2003 Cornelius et a1, Suite 1, Grass Valley, CA (US) 95945 ( * ) Notice: Subject to any disclaimer, the term of this patent is extended or adjusted under 35 OTHER PUBLICATIONS U.S.C. 154(b) by 199 days. _ Hannah V. Carey, The American Physiological Society, Physical _ Rev 83; Mammalian Hibernation: Cellular and Molecular (21) Appl' NO" 10/444’845 Responses to Depressed Metabolism and Low Temperature, 2003, (22) Filed: May 23, 2003 PP' 1153'1181' _ _ _ Primary ExamineriRuth A Davis (65) Pnor Pubhcatlon Data (74) Attorney, Agent, or F irmiBorson LaW Group, PC; D. US 2003/0228628 A1 Dec. 11, 2003 Benjamin Borson Related US. Application Data (57) ABSTRACT (60) Provisional application No. 60/382,913, ?led on May $112002’ provlslonal apphcanon NO‘ 60/383’271’ New methods for diagnosis of human dormancy syndrome e on May 24’ 2002' are provided. Human dormancy syndrome is characterized (51) Int C1 by elevated serum ratio of rT3/iT 3 compared to a population 1462K 3'9/00 (2006 01) of normal subjects from Which subjects suffering from A 61K 38/22 (200601) ?bromyalgia, chronic fatigue, obesity, dementias including A 61K 38/27 (200601) AlZheimer’s Disease and related dormancy conditions are C1 2 Q 1/00 (200601) excluded, and the presence of one or more ?ndings related ' A / A / A to reduced activity including torpor, chronic fatigue, insulin (52) US. -

Comprehensive Thyroid Plus Adrenal Report
Comprehensive Thyroid Plus Adrenal Report Jane Doe SAMPLEDate Collected: 2/13/2017 Comprehensive Thyroid Plus Adrenal Report Patient Name: Doe, Jane Batch Number: B0000 Patient DOB: 12/10/1960 Accession Number: Q00000 Gender: F Date Received: 2/14/2017 Physician Jon Doe, ND Report Date: 2/22/2017 Test Patient Results Reference Value DHEAS µg/dL 0 125 250 375 500 107 35 - 430 TSH µIU/mL 0.020 1.260 2.500 3.740 4.980+ 7.030 0.358 - 3.74 T4, Total µg/dL 0.5 4.4 8.3 12.1 16.0 7.2 4.5 - 12.5 T3, Free pg/mL 0.5 1.9 3.3 4.6 6.0 3.6 2.2 - 4.0 T4, Free ng/dL 0.10 0.83 1.55 2.28 3.00 0.87 0.76 - 1.46 Cortisol µg/dL 0.2 11.4 22.6 33.8 45.0 6.5 3.1 - 22.4 AntiTPO Ab IU/mL 10.0 20.0 30.0 40.0 50.0 + > 1000.0 0.0 - 35.0 AntiThyroglobulin Ab IU/mL 20 30 40 50 60 + 97 ND - 40 Thyroglobulin ng/mL 0 20 40 60 80 38 <= 55 Thyroxinebinding globulin, TBG µg/mL SAMPLE0 11 23 34 45 20 14 - 31 10401 Town Park Drive, Houston, Texas 77072 USA CLIA# 45D0710715 (800)227-LABS(5227) / (713)-621-3101 James W. Jacobson, Ph.D., Laboratory Director Comprehensive Thyroid Plus Adrenal Report Patient Name: Doe, Jane Batch Number: B0000 Patient DOB: 12/10/1960 Accession Number: Q00000 Gender: F Date Received: 2/14/2017 Physician Jon Doe, ND Report Date: 2/22/2017 Test Component Flag Result Reference Range DHEA-S µg/dL 107 35 - 430 TSH µIU/mL H 7.030 0.358 - 3.74 T4, Total µg/dL 7.2 4.5 - 12.5 T3, Free pg/mL 3.6 2.2 - 4.0 T4, Free ng/dL 0.87 0.76 - 1.46 Cortisol µg/dL 6.5 3.1 - 22.4 Anti-TPO Ab IU/mL H > 1000.0 0.0 - 35.0 Anti-Thyroglobulin Ab IU/mL H 97 ND - 40 Thyroglobulin ng/mL 38 <= 55 Thyroxine-binding globulin, TBGSAMPLE µg/mL 20 14 - 31 10401 Town Park Drive, Houston, Texas 77072 USA CLIA# 45D0710715 (800)227-LABS(5227) / (713)-621-3101 James W. -

Chang Et Al Thyroid
Available online at www.sciencedirect.com Toxicology 243 (2008) 330–339 Thyroid hormone status and pituitary function in adult rats given oral doses of perfluorooctanesulfonate (PFOS)ଝ Shu-Ching Chang a, Julie R. Thibodeaux b,1, Mary L. Eastvold c, David J. Ehresman a, James A. Bjork d, John W. Froehlich e,2, Christopher Lau b, Ravinder J. Singh c, Kendall B. Wallace d, John L. Butenhoff a,∗ a Medical Department, 3M Company, St. Paul, MN 55144, United States b United States Environmental Protection Agency, ORD, NHEERL, Reproductive Toxicology Division, Research Triangle Park, NC 27711, United States c Mayo Clinic and Foundation, Department of Laboratory Medicine and Pathology, Rochester, MN 55095, United States d University of Minnesota, Medical School, Department of Biochemistry and Molecular Biology, Duluth, MN 55812, United States e Pace Analytical Services, Inc., Minneapolis, MN 55414, United States Received 29 August 2007; received in revised form 18 October 2007; accepted 20 October 2007 Available online 26 October 2007 Abstract Introduction: Perfluorooctanesulfonate (PFOS) is widely distributed and persistent in humans and wildlife. Prior toxicological studies have reported decreased total and free thyroid hormones in serum without a major compensatory rise in thyrotropin (TSH) or altered thyroid gland histology. Although these animals (rats, mice and monkeys) might have maintained an euthyroid state, the basis for hypothyroxinemia remained unclear. We undertook this study to investigate the causes for the PFOS-induced reduction of serum total thyroxine (TT4) in rats. Hypotheses: We hypothesized that exposure to PFOS may increase free thyroxine (FT4) in the rat serum due to the ability of PFOS to compete with thyroxine for binding proteins. -

Ontogenesis of Iodothyronine-5'-Deiodinase
Ontogenesis of iodothyronine-5'-deiodinase. Induction of 5'- deiodinating activity by insulin, glucocorticoid, and thyroxine in cultured fetal mouse liver. K Sato, … , T Tsushima, K Shizume J Clin Invest. 1984;74(6):2254-2262. https://doi.org/10.1172/JCI111652. Research Article To elucidate the regulatory mechanism of ontogenetic development of iodothyronine-5'-deiodinase in the fetal and neonatal period, fetal mouse liver of the 19th day of gestation, in which no iodothyronine-5'-deiodinating activity was detectable, was cultured in Dulbecco-Vogt medium supplemented with 10% thyroid hormone-depleted fetal calf serum, insulin, hydrocortisone, and thyroid hormones. Iodothyronine-5'-deiodinating activity of the homogenate was assessed by the amount of iodide released from outer-ring-labeled reverse T3 and expressed as picomoles of 127I- per milligram of protein per minute. The enzyme activity was induced in a dose-dependent manner; optimal concentrations for insulin, hydrocortisone, and thyroxine were 1 microgram/ml, 0.4 microgram/ml, and 10(-6) M, respectively. Without supplementation of either hydrocortisone or thyroxine, no 5'-deiodination was detected. The enzyme activity was observed after 3 d of culture, peaked at days 14-20, and then gradually decreased. Lineweaver-Burk analysis revealed that the increase in activity was primarily due to an increase in Vmax (day 3, 0.2 pmol/mg protein per min; day 20, 2.5 pmol/mg protein per min). Half maximal thyroxine (T4) and triiodothyronine (T3) concentrations were 1 X 10(-7) M (free T4: 4 X 10(-10) M), and 2 X 10(-9) M (free T3: 5.0 X 10(-11) M), respectively, whereas reverse T3 did not elicit any activity at 10(-8)-10(-6) M. -

Dissertation / Doctoral Thesis
DISSERTATION / DOCTORAL THESIS Titel der Dissertation /Title of the Doctoral Thesis „ The natural compound curcumin: impact of cellular uptake and metabolism on in vitro activity “ verfasst von / submitted by Qurratul Ain Jamil angestrebter akademischer Grad / in partial fulfilment of the requirements for the degree of Doktorin der Naturwissenschaften (Dr.rer.nat.) Wien, 2018 / Vienna 2018 Studienkennzahl lt. Studienblatt / A 796 610 449 degree programme code as it appears on the student record sheet: Dissertationsgebiet lt. Studienblatt / Pharmazie, Klinische Pharmazie und Diagnostik field of study as it appears on the student record sheet: Pharmacy, Clinical Pharmacy and Diagnostics Betreut von / Supervisor: Ao.Univ.Prof.Magpharm.Dr.rer.nat.WalterJӓger “Verily in the Creation of the Heavens and the Earth, and in the Alteration of Night and Day, and the Ships which Sail through the Sea with that which is of used to Mankind, and the Water (Rain) which God sends down from the Sky and makes Earth Alive there with after its Death, and the moving Creatures of all kind that he has Scattered therein, and the Veering of Winds and Clouds which are held between the Sky and the Earth, are indeed Signs for Peoples of Understanding.” Acknowledgements While writing this section, I remember the first day in my lab, having a warm welcome from my supervisor; ao. Univ.-Prof. Mag. Dr. Walter Jӓger (Division of Clinical Pharmacy and Diagnostics, University of Vienna). He was waiting for me, offered me coffee and told me how to operate the coffee machine. He showed me, my office and asked me to make a list of all stuff, I need for daily work. -

A Bioinformatic and Mechanistic Study Elicits The
Journal name: Drug Design, Development and Therapy Article Designation: Original Research Year: 2015 Volume: 9 Drug Design, Development and Therapy Dovepress Running head verso: He et al Running head recto: UA reverses liver fibrosis in rats open access to scientific and medical research DOI: http://dx.doi.org/10.2147/DDDT.S85426 Open Access Full Text Article ORIGINAL RESEARCH A bioinformatic and mechanistic study elicits the antifibrotic effect of ursolic acid through the attenuation of oxidative stress with the involvement of ERK, PI3K/Akt, and p38 MAPK signaling pathways in human hepatic stellate cells and rat liver Wenhua He1,* Abstract: NADPH oxidases (NOXs) are a predominant mediator of redox homeostasis in Feng Shi1,* hepatic stellate cells (HSCs), and oxidative stress plays an important role in the pathogenesis Zhi-Wei Zhou2,* of liver fibrosis. Ursolic acid (UA) is a pentacyclic triterpenoid with various pharmacological Bimin Li1 activities, but the molecular targets and underlying mechanisms for its antifibrotic effect in the Kunhe Zhang1 liver remain elusive. This study aimed to computationally predict the molecular interactome Xinhua Zhang1 and mechanistically investigate the antifibrotic effect of UA on oxidative stress, with a focus on NOX4 activity and cross-linked signaling pathways in human HSCs and rat liver. Drug–drug Canhui Ouyang1 interaction via chemical–protein interactome tool, a server that can predict drug–drug interac- Shu-Feng Zhou2 tion via chemical–protein interactome, was used to predict the molecular targets of UA, and 1 Xuan Zhu Database for Annotation, Visualization, and Integrated Discovery was employed to analyze 1Department of Gastroenterology, the the signaling pathways of the predicted targets of UA. -

Thyroid Hormone
THYROID HORMONE Edited by Neeraj Kumar Agrawal Thyroid Hormone http://dx.doi.org/10.5772/2964 Edited by Neeraj Kumar Agrawal Contributors Pradip K. Sarkar, Asano Ishikawa, Jun Kitano, José María Fernández-Santos, Jesús Morillo- Bernal, Rocío García-Marín, José Carmelo Utrilla, Inés Martín-Lacave, Irmgard D. Dietzel, Sivaraj Mohanasundaram, Vanessa Niederkinkhaus, Gerd Hoffmann, Jens W. Meyer, Christoph Reiners, Christiana Blasl, Katharina Bohr, R.G. Ahmed, N.K. Agrawal, Ved Prakash, Manuj Sharma, Giuseppe Pasqualetti, Angela Dardano, Sara Tognini, Antonio Polini, Fabio Monzani, Renata de Azevedo Melo Luvizotto, Sandro José Conde, Miriane de Oliveira, Maria Teresa De Sibio, Keize Nagamati Jr, Célia Regina Nogueira, Eva Feigerlova, Marc Klein, Anna Angelousi, Lelia Groza, Bruno Leheup, Georges Weryha, Einav Yehuda-Shnaidman, Bella Kalderon, Jacob Bar-Tana, Emina Kasumagic-Halilovic, Begler Begovic, Francesco Torino, Agnese Barnabei, Roberto Baldelli, Marialuisa Appetecchia, Clara Spinel, Magnolia Herrera Published by InTech Janeza Trdine 9, 51000 Rijeka, Croatia Copyright © 2012 InTech All chapters are Open Access distributed under the Creative Commons Attribution 3.0 license, which allows users to download, copy and build upon published articles even for commercial purposes, as long as the author and publisher are properly credited, which ensures maximum dissemination and a wider impact of our publications. After this work has been published by InTech, authors have the right to republish it, in whole or part, in any publication of which they are the author, and to make other personal use of the work. Any republication, referencing or personal use of the work must explicitly identify the original source. Notice Statements and opinions expressed in the chapters are these of the individual contributors and not necessarily those of the editors or publisher. -

Expression and Function of Organic Anion Transporting Polypeptides in the Human Brain: Physiological and Pharmacological Implications
pharmaceutics Review Expression and Function of Organic Anion Transporting Polypeptides in the Human Brain: Physiological and Pharmacological Implications Anima M. Schäfer 1, Henriette E. Meyer zu Schwabedissen 1 and Markus Grube 2,* 1 Biopharmacy, Department Pharmaceutical Sciences, University of Basel, Klingelbergstrasse 50, 4056 Basel, Switzerland; [email protected] (A.M.S.); [email protected] (H.E.M.z.S.) 2 Center of Drug Absorption and Transport (C_DAT), Department of Pharmacology, University Medicine of Greifswald, 17489 Greifswald, Germany * Correspondence: [email protected]; Tel./Fax: +49-3834-865636 Abstract: The central nervous system (CNS) is an important pharmacological target, but it is very effectively protected by the blood–brain barrier (BBB), thereby impairing the efficacy of many potential active compounds as they are unable to cross this barrier. Among others, membranous efflux transporters like P-Glycoprotein are involved in the integrity of this barrier. In addition to these, however, uptake transporters have also been found to selectively uptake certain compounds into the CNS. These transporters are localized in the BBB as well as in neurons or in the choroid plexus. Among them, from a pharmacological point of view, representatives of the organic anion transporting polypeptides (OATPs) are of particular interest, as they mediate the cellular entry of Citation: Schäfer, A.M.; Meyer zu a variety of different pharmaceutical compounds. Thus, OATPs in the BBB potentially offer the Schwabedissen, H.E.; Grube, M. possibility of CNS targeting approaches. For these purposes, a profound understanding of the Expression and Function of Organic expression and localization of these transporters is crucial. -

Clinical Applications
HORMONE BALANCE CLINICAL APPLICATIONS © 2014 SpectraCell Laboratories, Inc. All rights reserved. DOC 502 12.14 Visit us at www.spectracell.com or call us at 800.227.LABS (5227) HORMONES 101 Steroid Hormones • Dehydroepiandrosterone sulfate (DHEAS) • Androstenedione • Testosterone • Estradiol (E2) • Estrone (E1) • Estriol (E3) • Progesterone Peptide Hormones • Sex hormone binding globulin (SHBG) • Luteinizing hormone (LH) • Follicle stimulating hormone (FSH) • Prolactin Hormone Biosynthesis Pathway DHEAS → Androstenedione → Testosterone or Estrogens Dehydroepiandrosterone sulfate (DHEAS) DHEA is the most abundant sex hormone in the body, with levels typically around 20 times that of any other steroid hormone. Produced primarily in the adrenal gland, it circulates the body in the form of DHEAS (sulfated form), and it is the major precursor hormone to androstenedione and subsequently estrogen and testosterone. DHEA has important functions unrelated to its role as a precursor to other sex hormones such as enhancing immunity to viruses (by increasing natural killer cell activity), alleviating autoimmune conditions, cancer prevention, bone health, cognitive function (via its role in the calming neurotransmitter GABA) and improving insulin sensitivity (by inhibiting glucose-6-phosphate dehydrogenase, the enzyme responsible for fat accumulation). Levels of DHEAS, which is synthesized through a series of reactions from cholesterol, peak when a person is in their mid 20’s and gradually decline in later decades of life. Increasing DHEAS levels may eventually increase levels of downstream sex hormones (testosterone and estrogen, but not necessarily progesterone) and the clinical effects are similar. In addition, the effects of DHEAS may depend on the levels of other hormones. For example, in premenopausal women, DHEAS seems to have a protective effect against hormone related cancers (breast, ovarian, uterine), while in postmenopausal women, if estrogen levels are low, DHEAS may contribute to growth of rogue cells in hormone sensitive tissues. -
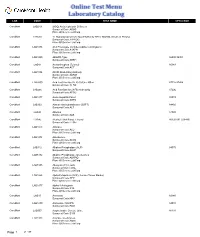
Antistreptolysin O Screen Sunquest Code:ASOS Place of Service:Labcorp Caromont LAB530 17 Hydroxprogesteron
LAB CODE TEST NAME CPT4 CODE CaroMont LAB219 (ASO) Antistreptolysin O Screen Sunquest Code:ASOS Place Of Service:LabCorp CaroMont LAB530 17 Hydroxprogesterone Quantitative by HPLC-MS/MS, Serum or Plasma Sunquest Code:17PROG Place Of Service:LabCorp CaroMont LAB1078 A1A Phenotype (Includes Alpha-1-Antitrypsin) Sunquest Code:A1APH Place Of Service:LabCorp CaroMont LAB1000 ABO/Rh Type 86900 86901 Sunquest Code:ABRH CaroMont LAB43 Acetaminophen (Tylenol) 80143 Sunquest Code:ACT CaroMont LAB1054 ACHR Modulating Antibody Sunquest Code:ACRM Place Of Service:LabCorp CaroMont LAB5000 Acid Fast Bacillus (AFB) Culture: Other 87116 87206 Sunquest Code:AFBO CaroMont LAB266 Acid Fast Bacillus (AFB) Stain Only 87206 Sunquest Code:AFSO CaroMont LAB1877 Acute Hepatitis Panel 80074 Sunquest Code:AHPA CaroMont LAB132 Alanine Aminotransferase (SGPT) 84460 Sunquest Code:ALT CaroMont LAB45 Albumin 82040 Sunquest Code:ALB CaroMont LAB46 Alcohol, Ethyl Blood, Ethanol 80320 001 (G0480) Sunquest Code:ETOH CaroMont LAB1073 Aldolase Sunquest Code:ALD Place Of Service:LabCorp CaroMont LAB1074 Aldosterone Sunquest Code:ALDS Place Of Service:LabCorp CaroMont LAB112 Alkaline Phosphatase (ALP) 84075 Sunquest Code:ALKP CaroMont LAB5132 Alkaline Phosphatase Isoenzymes Sunquest Code:ALKPIO Place Of Service:LabCorp CaroMont LAB5105 Allergens (19) Foods Sunquest Code:FDAL Place Of Service:LabCorp CaroMont LAB1068 Alpha Fetoprotein (AFP), Serum (Tumor Marker) Sunquest Code:AFP Place Of Service:LabCorp CaroMont LAB1077 Alpha-1-Antitrypsin Sunquest Code:A1A Place Of Service:LabCorp -

The Peripheral Conversion of Thyroxine (T4) Into Triiodothyronine
The Peripheral Conversion of Thyroxine (T4) into i • Triiodothyronine (T3) and Reverse triiodothyronine (rT3) 3 The Peripheral Conversion of Thyroxine (T4) into Triiodothyronine (T3) sts- a», and Reverse triiodothyronine (rT3) ACADEMISCH PROEFSCHRIFT ter verkrijging van de graad van doctor in de Geneeskunde aan de Universiteit van Amsterdam, ?••!• op gezag van de Rector Magnificus dr. J.Bruyn, hoogleraar in de Faculteit der Letteren, in het openbaar te verdedigen in de aula der Universiteit (tijdelijk in de Lutherse Kerk, ingang Singel 411, hoek Spui), op donderdag 15 november 1979 om 16.00 uur precies door WILLEM MAARTEN WIERSINGA geboren te Leiden AMSTERDAM 1979 I; Promoter : Dr. J.L. Touber g Coreferent : Prof. dr. A. Querido 'l Copromotor : Prof. dr. M. Koster Dit proefschrift is bewerkt op de afdeling endocrinologie (Dr. J.L. Touber) van de kliniek voor inwendige ziekten (Prof.Dr. M. Koster en Prof.Dr. J. Vreeken) van het Wilhelmina Gasthuis, Academisch Ziekenhuis bij de Universiteit van Amsterdam. Het verschijnen van dit proefschrift werd mede mogeiijk gemaakt door steun van de Nederlandse Hartstichting en van ICI Holland B.V. I i*' Ii: VOORWOORD I Dit proefschrift is niet opgedragen aan iemand in het bijzonder, daar het tv, verrichte onderzoek in eerste instantie mijn eigen nieuwsgierigheid heeft 1} bevredigd en ik de indruk heb dat het meer mijn eigen genoegen heeft r ' gediend dan dat van anderen. Een woord van dank aan de velen die in de : afgelopen jaren bijgedragen hebben tot deze studies, is wel op zijn plaats, '. daar het onderzoek zonder hun steun niet mogelijk zou zijn geweest, en ; vooral ook omdat ik met plezier terugdenk aan de onderlinge , samenwerking.