Seed Total Protein Profiling in Discrimination of Closely Related Pines: Evidence from the Pinus Mugo Complex
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Potential Impact of Climate Change
Adhikari et al. Journal of Ecology and Environment (2018) 42:36 Journal of Ecology https://doi.org/10.1186/s41610-018-0095-y and Environment RESEARCH Open Access Potential impact of climate change on the species richness of subalpine plant species in the mountain national parks of South Korea Pradeep Adhikari, Man-Seok Shin, Ja-Young Jeon, Hyun Woo Kim, Seungbum Hong and Changwan Seo* Abstract Background: Subalpine ecosystems at high altitudes and latitudes are particularly sensitive to climate change. In South Korea, the prediction of the species richness of subalpine plant species under future climate change is not well studied. Thus, this study aims to assess the potential impact of climate change on species richness of subalpine plant species (14 species) in the 17 mountain national parks (MNPs) of South Korea under climate change scenarios’ representative concentration pathways (RCP) 4.5 and RCP 8.5 using maximum entropy (MaxEnt) and Migclim for the years 2050 and 2070. Results: Altogether, 723 species occurrence points of 14 species and six selected variables were used in modeling. The models developed for all species showed excellent performance (AUC > 0.89 and TSS > 0.70). The results predicted a significant loss of species richness in all MNPs. Under RCP 4.5, the range of reduction was predicted to be 15.38–94.02% by 2050 and 21.42–96.64% by 2070. Similarly, under RCP 8.5, it will decline 15.38–97.9% by 2050 and 23.07–100% by 2070. The reduction was relatively high in the MNPs located in the central regions (Songnisan and Gyeryongsan), eastern region (Juwangsan), and southern regions (Mudeungsan, Wolchulsan, Hallasan, and Jirisan) compared to the northern and northeastern regions (Odaesan, Seoraksan, Chiaksan, and Taebaeksan). -
IUCN Red List of Threatened Species™ to Identify the Level of Threat to Plants
Ex-Situ Conservation at Scott Arboretum Public gardens and arboreta are more than just pretty places. They serve as an insurance policy for the future through their well managed ex situ collections. Ex situ conservation focuses on safeguarding species by keeping them in places such as seed banks or living collections. In situ means "on site", so in situ conservation is the conservation of species diversity within normal and natural habitats and ecosystems. The Scott Arboretum is a member of Botanical Gardens Conservation International (BGCI), which works with botanic gardens around the world and other conservation partners to secure plant diversity for the benefit of people and the planet. The aim of BGCI is to ensure that threatened species are secure in botanic garden collections as an insurance policy against loss in the wild. Their work encompasses supporting botanic garden development where this is needed and addressing capacity building needs. They support ex situ conservation for priority species, with a focus on linking ex situ conservation with species conservation in natural habitats and they work with botanic gardens on the development and implementation of habitat restoration and education projects. BGCI uses the IUCN Red List of Threatened Species™ to identify the level of threat to plants. In-depth analyses of the data contained in the IUCN, the International Union for Conservation of Nature, Red List are published periodically (usually at least once every four years). The results from the analysis of the data contained in the 2008 update of the IUCN Red List are published in The 2008 Review of the IUCN Red List of Threatened Species; see www.iucn.org/redlist for further details. -

Evolution of the Female Conifer Cone Fossils, Morphology and Phylogenetics
DEPARTMENT OF BIOLOGICAL AND ENVIRONMENTAL SCIENCES EVOLUTION OF THE FEMALE CONIFER CONE FOSSILS, MORPHOLOGY AND PHYLOGENETICS Daniel Bäck Degree project for Bachelor of Science with a major in Biology BIO602, Biologi: Examensarbete – kandidatexamen, 15 hp First cycle Semester/year: Spring 2020 Supervisor: Åslög Dahl, Department of Biological and Environmental Sciences Examiner: Claes Persson, Department of Biological and Environmental Sciences Front page: Abies koreana (immature seed cones), Gothenburg Botanical Garden, Sweden Table of contents 1 Abstract ............................................................................................................................... 2 2 Introduction ......................................................................................................................... 3 2.1 Brief history of Florin’s research ............................................................................... 3 2.2 Progress in conifer phylogenetics .............................................................................. 4 3 Aims .................................................................................................................................... 4 4 Materials and Methods ........................................................................................................ 4 4.1 Literature: ................................................................................................................... 4 4.2 RStudio: ..................................................................................................................... -
Botanical Name Common Name
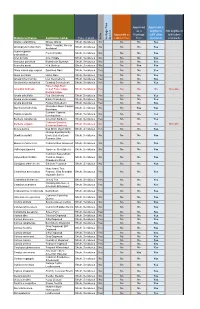
Approved Approved & as a eligible to Not eligible to Approved as Frontage fulfill other fulfill other Type of plant a Street Tree Tree standards standards Heritage Tree Tree Heritage Species Botanical Name Common name Native Abelia x grandiflora Glossy Abelia Shrub, Deciduous No No No Yes White Forsytha; Korean Abeliophyllum distichum Shrub, Deciduous No No No Yes Abelialeaf Acanthropanax Fiveleaf Aralia Shrub, Deciduous No No No Yes sieboldianus Acer ginnala Amur Maple Shrub, Deciduous No No No Yes Aesculus parviflora Bottlebrush Buckeye Shrub, Deciduous No No No Yes Aesculus pavia Red Buckeye Shrub, Deciduous No No Yes Yes Alnus incana ssp. rugosa Speckled Alder Shrub, Deciduous Yes No No Yes Alnus serrulata Hazel Alder Shrub, Deciduous Yes No No Yes Amelanchier humilis Low Serviceberry Shrub, Deciduous Yes No No Yes Amelanchier stolonifera Running Serviceberry Shrub, Deciduous Yes No No Yes False Indigo Bush; Amorpha fruticosa Desert False Indigo; Shrub, Deciduous Yes No No No Not eligible Bastard Indigo Aronia arbutifolia Red Chokeberry Shrub, Deciduous Yes No No Yes Aronia melanocarpa Black Chokeberry Shrub, Deciduous Yes No No Yes Aronia prunifolia Purple Chokeberry Shrub, Deciduous Yes No No Yes Groundsel-Bush; Eastern Baccharis halimifolia Shrub, Deciduous No No Yes Yes Baccharis Summer Cypress; Bassia scoparia Shrub, Deciduous No No No Yes Burning-Bush Berberis canadensis American Barberry Shrub, Deciduous Yes No No Yes Common Barberry; Berberis vulgaris Shrub, Deciduous No No No No Not eligible European Barberry Betula pumila -

Plant in the Spotlight
TheThe AmericanAmerican GARDENERGARDENER® TheThe MagazineMagazine ofof thethe AAmericanmerican HorticulturalHorticultural SocietySociety March / April 2010 Beautiful, Durable Baptisias Coniferous Groundcovers DynamicDynamic DuetsDuets Agaves for Small Spaces forfor ShadeShade contents Volume 89, Number 2 . March / April 2010 FEATURES DEPARTMENTS 5 NOTES FROM RIVER FARM 6 MEMBERS’ FORUM 8 NEWS FROM AHS Allan Armitage to host AHS webinar, River Farm Spring Garden Market in April, AHS National Children & Youth Garden Symposium goes to California, AHS to participate in 4th annual Washington, D.C.-area Garden Fest, 2010 AHS President’s Council Members Trip to Florida. 14 AHS NEWS SPECIAL 2010 Great American Gardeners National Award winners and 2010 Book Award winners. 42 ONE ON ONE WITH… page 36 Steven Still: Herbaceous perennial expert. 44 HOMEGROWN HARVEST A bumper crop of broccoli. 18 DYNAMIC DUETS FOR SHADE BY KRIS WETHERBEE Light up shady areas of the garden by using plant combinations 46 GARDENER’S NOTEBOOK that offer complementary textures and colors. Mt. Cuba Center releases coneflower evaluation results, AMERICAN BEAUTIES: study shows bumble bee 24 page 24 populations declining, BAPTISIAS BY RICHARD HAWKE GreatPlants® and Perennial The release of new cultivars of Plant Association name 2010 false indigo has renewed garden- Plants of the Year, Berry ers’ interest in the genus Baptisia. Botanic Garden to close, Jane Pepper retires as president of page 46 GROUND-COVERING Pennsylvania Horticultural 30 Society. CONIFERS BY PENELOPE O’SULLIVAN 50 GREEN GARAGE® Reduce maintenance and add Garden gloves. vibrant color and texture to the garden by using low-growing 52 BOOK REVIEWS conifers as groundcovers. What’s Wrong with My Plant? (And How Do I Fix It?); Homegrown Vegetables, Fruits, and Herbs; The Vegetable Gardener’s Bible; and 36 AGAVES FOR SMALL GARDENS BY MARY IRISH The Encyclopedia of Herbs. -

Specializing in Rare and Unique Trees 2021 Catalogue Pot Sizes: the Number Represents the Size of the Pot Ie
Whistling Gardens Ltd., 698 Concession 3, Wilsonville, ON N0E 1Z0 Phone: 519-443-5773 Fax: 519-443-4141 Email: [email protected] Specializing in Rare and Unique Trees 2021 Catalogue Pot sizes: The number represents the size of the pot ie. #1= 1 gallon, #10 = 10 gallon #1 potted conifers are usually 3-5years old. #10 potted conifers dwarf conifers are between 10 and 15 years old #1 trees= usually seedlings #10 trees= can be several years old anywhere from 5 to 10' tall depending on species and variety. Please ask us on sizes and varieties you are not sure about. Many plants are limited to 1 specimen. To reserve your plant(s) a 25% is required. Plants should be picked up by June 15th. Most plants arrive at the gardens by May 10th. Guarantee: We cannot control the weather (good or bad), rodents (big or small), pests (teenie, tiny), poor siting, soil types, lawnmovers, snowplows etc. Plants we carry are expected to grow within the parameters of normal weather conditons. All woody plant purchases are guaranteed from time of purchase to December 1st of current year. Perennials are not guaranteed. Any plant not performing or dying in current season will be happily replaced or credited towards a new plant. CategoryOrderedAvailable Plant Size Price Description C Fir C 2 Abies concolor Archer's Dwarf #5 $ 145.00 C 1 Abies concolor Candicans #3 $ 120.00 C 1 Abies concolor Wintergold #7 $ 240.00 Light green in summer, bright yellow in winter. Slower growing. Korean Fir C 2 Abies koreana Aurea #5 $ 180.00 Lemon yellow foliage year round. -

The Firs by Dr
The Firs by Dr. Bert Cregg s a native of the Pacific limiting factor. We can adjust soil pH MSU Project GREEEN, we have Northwest, I have always had by applying sulfur or fertilizing with transplanted over 200 of the trees to Aa soft spot in my heart for firs. ammonium sulfate to make a site MSU Horticulture Research Stations From high school summers shearing more suitable for firs, whereas soil across the state and are expanding the deep green grand firs (Abies grandis) drainage is difficult to change across evaluation to include traits that are on Christmas tree plantations to a landscape. Nevertheless, there are critical in landscapes such as response seeing the spire-like crowns of several reasons to expect the use of to soil pH and drought. subalpine fir Abies( lasiocarpa) on firs to increase in the future: day trips to Mt. Rainier, firs were an 3. Introduction of Hybrids ever-present symbol of my Northwest 1. Blue Spruce Burnout One of the interesting features of upbringing. The true firs (genus Since I’ve started this column firs is that many species hybridize Abies) include a remarkable variety people have asked me, “Why do readily. It may be possible through of trees that are important to the you hate blue spruce so much?” hybridization to combine desirable nursery, landscape, and Christmas For the record, I don’t hate blue ornamental traits from one species tree industries of Michigan. spruce (or Norway spruce or Scots (say, needle color or form) with The genus includes over 40 pine or Austrian pine or white pine). -

Whitehallhouse & GARDENS
HOUSE & Whitehall GARDENS SPECIMEN GARDEN PLANT LIST SUN | PART SHADE | SHADE | BLOOM TIME Abelia x grandiflora Abies koreana ‘Horstmann’s Silberlocke’ Albizia julibrissin ‘Ishii Weeping’ Glossy abelia Horstmann’s Silberlocke Korean fir Weeping mimosa FAMILY: Caprifoliaceae FAMILY: Pinaceae FAMILY: Fabaceae PLANT TYPE: Deciduous shrub TREE TYPE: Needled evergreen TREE TYPE: Deciduous | June to September | Nonflowering | June and July Amsonia hubrichtii Anemone hupehensis Asclepias tuberosa Blue star Japanese thimbleweed Butterfly weed FAMILY: Apocynaceae FAMILY: Ranunculaceae FAMILY: Apocynaceae PLANT TYPE: Perennial PLANT TYPE: Perennial PLANT TYPE: Perennial | April to May | August and September | June and July Buddleja davidii Buxus sempervirens ‘Dee Runk’ Campanula ‘Sarastro’ Butterfly bush Dee Runk boxwood Bellflower FAMILY: Scrophulariaceae FAMILY: Buxaceae FAMILY: Campanulaceae PLANT TYPE: Deciduous shrub TREE TYPE: Broadleaf evergreen PLANT TYPE: Perennial | June to September | Nonflowering | June to August 1 HOUSE & Whitehall GARDENS SPECIMEN GARDEN PLANT LIST SUN | PART SHADE | SHADE | BLOOM TIME Capinus betulus ‘Columnaris Nana’ Chaenomeles speciose ‘Double Take Scarlet’ Chianthus retusus ‘Ivory Tower’ Dwarf European hornbeam Double Take flowering quince Chinese fringe tree FAMILY: Betulaceae FAMILY: Rosaceae FAMILY: Oleaceae TREE TYPE: Deciduous PLANT TYPE: Deciduous shrub TREE TYPE: Deciduous | March | March and April | May and June Chionanthus pygmaeus Chionanthus retusus ‘China Snow’ Chrysanthemum Dwarf pygmy fringe tree -

Korean Red List of Threatened Species Korean Red List Second Edition of Threatened Species Second Edition Korean Red List of Threatened Species Second Edition
Korean Red List Government Publications Registration Number : 11-1480592-000718-01 of Threatened Species Korean Red List of Threatened Species Korean Red List Second Edition of Threatened Species Second Edition Korean Red List of Threatened Species Second Edition 2014 NIBR National Institute of Biological Resources Publisher : National Institute of Biological Resources Editor in President : Sang-Bae Kim Edited by : Min-Hwan Suh, Byoung-Yoon Lee, Seung Tae Kim, Chan-Ho Park, Hyun-Kyoung Oh, Hee-Young Kim, Joon-Ho Lee, Sue Yeon Lee Copyright @ National Institute of Biological Resources, 2014. All rights reserved, First published August 2014 Printed by Jisungsa Government Publications Registration Number : 11-1480592-000718-01 ISBN Number : 9788968111037 93400 Korean Red List of Threatened Species Second Edition 2014 Regional Red List Committee in Korea Co-chair of the Committee Dr. Suh, Young Bae, Seoul National University Dr. Kim, Yong Jin, National Institute of Biological Resources Members of the Committee Dr. Bae, Yeon Jae, Korea University Dr. Bang, In-Chul, Soonchunhyang University Dr. Chae, Byung Soo, National Park Research Institute Dr. Cho, Sam-Rae, Kongju National University Dr. Cho, Young Bok, National History Museum of Hannam University Dr. Choi, Kee-Ryong, University of Ulsan Dr. Choi, Kwang Sik, Jeju National University Dr. Choi, Sei-Woong, Mokpo National University Dr. Choi, Young Gun, Yeongwol Cave Eco-Museum Ms. Chung, Sun Hwa, Ministry of Environment Dr. Hahn, Sang-Hun, National Institute of Biological Resourses Dr. Han, Ho-Yeon, Yonsei University Dr. Kim, Hyung Seop, Gangneung-Wonju National University Dr. Kim, Jong-Bum, Korea-PacificAmphibians-Reptiles Institute Dr. Kim, Seung-Tae, Seoul National University Dr. -

Bulletin, Volume 53 (1993)
Tree-Ring Bulletin, Volume 53 (1993) Item Type Article Publisher Tree-Ring Society Journal Tree-Ring Bulletin Rights Copyright © Tree-Ring Society. All rights reserved. Download date 28/09/2021 15:30:30 Link to Item http://hdl.handle.net/10150/263019 TREE -RING BULLETIN 1993 PUBLISHED BY THE TREE RING SOCIETY with the cooperation of THE LABORATORY OF TREE -RING RESEARCH THE UNIVERSITY OF ARIZONA® Printed in 1995 TREE -RING BULLETIN EDITORIAL POLICY The Tree -Ring Bulletin is devoted to papers dealing with the growth rings of trees, and the application of tree -ring studies to problems in a wide variety of fields, including but not limit- ed to archaeology, geology, ecology, hydrology, climatology, forestry, and botany. Papers involving research results, new techniques of data acquisition or analysis, and regional or sub- ject oriented reviews or syntheses are considered for publication. Two categories of manuscripts are considered. Articles should not exceed 5000 words, or approximately 20 double- spaced typewritten pages, including tables, references, and an abstract of 200 words or fewer. All manuscripts submitted as Articles are reviewed by at least two referees.Research Reports, which normally are not reviewed, should not exceed 1500 words or include more than two figures. Research Reports address technical developments, describe well- documented but preliminary research results, or present findings for which the Article format is not appropriate. Papers are published only in English, but abstracts of Articles appear in at least two addi- tional languages. Contributors are encouraged to include German and/or French translations of the abstracts with their manuscripts. Abstracts in other languages may be printed if sup- plied by the author(s) with English translations. -

Wood Anatomy of the Genus Abies Luis García Esteban*, Paloma
IAWA Journal, Vol. 30 (3), 2009: 231–245 WOOD ANATOMY OF THE GENUS ABIES A REVIEW Luis García Esteban*, Paloma de Palacios, Francisco García Fernández and Ruth Moreno Universidad Politécnica de Madrid. Escuela Técnica Superior de Ingenieros de Montes, Departamento de Ingeniería Forestal, Ciudad Universitaria, 28040 Madrid, Spain *Corresponding author [E-mail: [email protected]] SUMMARY The literature on the wood anatomy of the genus Abies is reviewed and discussed, and complemented with a detailed study of 33 species, 1 sub- species and 4 varieties. In general, the species studied do not show diag- nostic interspecific differences, although it is possible to establish differences between groups of species using certain quantitative and quali- tative features. The marginal axial parenchyma consisting of single cells and the ray parenchyma cells with distinctly pitted horizontal walls, nodular end walls and presence of indentures are constant for the genus, although these features also occur in the other genera of the Abietoideae. The absence of ray tracheids in Abies can be used to distinguish it from Cedrus and Tsuga, and the irregularly shaped parenchymatous marginal ray cells are only shared with Cedrus. The absence of resin canals enables Abies to be distinguished from very closely related genera such as Keteleeria and Nothotsuga. The crystals in the ray cells, taxodioid cross-field pitting and the warty layer in the tracheids can be regarded as diagnostic generic features. Key words: Abies, Abietoideae, anatomy, wood. INTRODUCTION The family Pinaceae, with 11 genera and 225 species, is the largest conifer family. The genus Abies, with 48 species and 24 varieties, has the second highest number of species after the genus Pinus (Farjon 2001). -

List of Korean Evergreen Plants
APPENDIX 1 List of Korean evergreen plants Species No. Family Name Species Name 1 Piperaceae Piper kadzura 2 Chloranthaceae Sarcandra glabra 3 Myricaceae Myrica rubra 4 Fagaceae Castanopsis cuspidata val. sieboldii 5 Castanopsis cuspidata val. latifolia 6 Castanopsis cuspidata val. thunbergii 7 Cyclobalanopsis acuta 8 Cyclobalanopsis acuta form. subserra 9 Cyclobalanopsis gilva 10 Cyclobalanopsis glauca 11 Cyclobalanopsis myrsinaefolia 12 Cyclobalanopsis stenophylla 13 Cyclobalanopsis stenophylla val. latifolia 14 Moraceae Ficus erecta 15 Ficus erecta val. longepedunculata 16 Ficus erecta val. sieboldii 17 Ficus nipponca 18 Ficus pumila ( = stipulata) 19 Loranthaceae Hypear tanakae 20 Scurrula yadoriki 21 Viscum coloratum val. lutescens 22 Viscum coloratum form. rubroauranticum 23 Bifaria Bifaria japonica 24 Lardizabalaceae Stauntonia hexaphylla 25 Menispermaceae Stephania japonica 26 Illiaceae Illicium anisatum 27 Lauraceae Kadsura japonica 28 Cinnamomum camphora 29 Cinnamomum japonicum 30 Cinnamomum loureirii 31 Fiwa japonica 32 Izosta lancifolia 33 Machilus japonica 34 M achilus thunbergii 35 Machilus thunbergii var. obovata 36 Neolitsea aciculata 37 Neolitsea sericea 38 Pittosporaceae Pittmporum lobira 39 Hamamelidaceae Distylium racemosum var. latifolium 40 Distylium racemosum var. typicum 41 Rosaceae Raphiolepsis liukiuensis 42 Raphiolepsis obovata 43 Raphiolepsis ubellata 44 Rubus buergeri 185 186 45 Rutaceae Citrus aurantium 46 Citrus deliciosa 47 Citrus grandis 48 Citrus junos 49 Citrus kinokuni 50 Citrus medica var. sarcodactylus 51 Citrus natsudaidai 52 Citrus noblis 53 Citrus sinensis 54 Citrus unshiu 55 Zanthoxylum planispinum 56 Daphniphyllaceae Daphniphyllum glaucescens 57 Daphniphyllum macropodum 58 Buxaceae Buxus koreana 59 Buxus koreana var. elongata 60 Buxus koreana var. insularis 61 Buxus microphylla 62 AquifoJiaceae !lex comuta form. typica 63 !lex crenata var. microphylla 64 !lex integra var.