(Abies Koreana) Vulnerable to Climate Change
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Formation of Spatial Mosaic of Abies Nephrolepis (Pinaceae) Populations in Korean Pine- Broadleaved Forests in the South of Russian Far East
Rastitelnye Resursy. 53(4): 480—495, 2017 FORMATION OF SPATIAL MOSAIC OF ABIES NEPHROLEPIS (PINACEAE) POPULATIONS IN KOREAN PINE- BROADLEAVED FORESTS IN THE SOUTH OF RUSSIAN FAR EAST © T. Ya. Petrenko, *, 1, 2 A. M. Omelko, 1A. A. Zhmerenetsky, 1 O. N. Ukhvatkina,1 L. A. Sibirina1 1 Federal Scientific center of the East Asia terrestrial biodiversity FEB RAS, Vladivostok, Russia 2Far Eastern Federal University, Vladivostok, Russia *E-mail: [email protected] SUMMARY We studied structure and described formation of Abies nephrolepis (Trautv.) Maxim. population mosaic in Korean pine-broadleaved forest of the Sikhote-Alin mountain range in the south of Russian Far East. The study was performed on two permanent sample plots (1.5 ha and 10.5 ha) established in primary forest of Verhneussuriysky Research Station of the Federal Scientific center of the East Asia terrestrial biodiversity, FEB RAS. One of the permanent sample plots (10.5 ha) was specifically designed for studying tree population mosaic. It covers an area necessary for the analysis of population structure of the dominant tree species. To describe the population mosaics we use demographic approach that allows to consider specific features of plant ontogeny. It is established, that mosaic structure transforms from contagious (immature plants) to normal (generative plants) distribution. Mosaic of generative plants is formed at the time of transition from immature to virginal ontogenetic stage. Unlike mosaic of Picea ajanensis (Siebold et Zucc.) Carr., where plants continuously accumulated starting from virginal stage, mosaic of A. nephrolepis continues to thin out starting from immature stage. Thus, this species is characterized by R-strategy. -

EVERGREEN TREES for NEBRASKA Justin Evertson & Bob Henrickson
THE NEBRASKA STATEWIDE ARBORETUM PRESENTS EVERGREEN TREES FOR NEBRASKA Justin Evertson & Bob Henrickson. For more plant information, visit plantnebraska.org or retreenbraska.unl.edu Throughout much of the Great Plains, just a handful of species make up the majority of evergreens being planted. This makes them extremely vulnerable to challenges brought on by insects, extremes of weather, and diseases. Utilizing a variety of evergreen species results in a more diverse and resilient landscape that is more likely to survive whatever challenges come along. Geographic Adaptability: An E indicates plants suitable primarily to the Eastern half of the state while a W indicates plants that prefer the more arid environment of western Nebraska. All others are considered to be adaptable to most of Nebraska. Size Range: Expected average mature height x spread for Nebraska. Common & Proven Evergreen Trees 1. Arborvitae, Eastern ‐ Thuja occidentalis (E; narrow habit; vertically layered foliage; can be prone to ice storm damage; 20‐25’x 5‐15’; cultivars include ‘Techny’ and ‘Hetz Wintergreen’) 2. Arborvitae, Western ‐ Thuja plicata (E; similar to eastern Arborvitae but not as hardy; 25‐40’x 10‐20; ‘Green Giant’ is a common, fast growing hybrid growing to 60’ tall) 3. Douglasfir (Rocky Mountain) ‐ Pseudotsuga menziesii var. glauca (soft blue‐green needles; cones have distinctive turkey‐foot bract; graceful habit; avoid open sites; 50’x 30’) 4. Fir, Balsam ‐ Abies balsamea (E; narrow habit; balsam fragrance; avoid open, windswept sites; 45’x 20’) 5. Fir, Canaan ‐ Abies balsamea var. phanerolepis (E; similar to balsam fir; common Christmas tree; becoming popular as a landscape tree; very graceful; 45’x 20’) 6. -

Potential Impact of Climate Change
Adhikari et al. Journal of Ecology and Environment (2018) 42:36 Journal of Ecology https://doi.org/10.1186/s41610-018-0095-y and Environment RESEARCH Open Access Potential impact of climate change on the species richness of subalpine plant species in the mountain national parks of South Korea Pradeep Adhikari, Man-Seok Shin, Ja-Young Jeon, Hyun Woo Kim, Seungbum Hong and Changwan Seo* Abstract Background: Subalpine ecosystems at high altitudes and latitudes are particularly sensitive to climate change. In South Korea, the prediction of the species richness of subalpine plant species under future climate change is not well studied. Thus, this study aims to assess the potential impact of climate change on species richness of subalpine plant species (14 species) in the 17 mountain national parks (MNPs) of South Korea under climate change scenarios’ representative concentration pathways (RCP) 4.5 and RCP 8.5 using maximum entropy (MaxEnt) and Migclim for the years 2050 and 2070. Results: Altogether, 723 species occurrence points of 14 species and six selected variables were used in modeling. The models developed for all species showed excellent performance (AUC > 0.89 and TSS > 0.70). The results predicted a significant loss of species richness in all MNPs. Under RCP 4.5, the range of reduction was predicted to be 15.38–94.02% by 2050 and 21.42–96.64% by 2070. Similarly, under RCP 8.5, it will decline 15.38–97.9% by 2050 and 23.07–100% by 2070. The reduction was relatively high in the MNPs located in the central regions (Songnisan and Gyeryongsan), eastern region (Juwangsan), and southern regions (Mudeungsan, Wolchulsan, Hallasan, and Jirisan) compared to the northern and northeastern regions (Odaesan, Seoraksan, Chiaksan, and Taebaeksan). -
IUCN Red List of Threatened Species™ to Identify the Level of Threat to Plants
Ex-Situ Conservation at Scott Arboretum Public gardens and arboreta are more than just pretty places. They serve as an insurance policy for the future through their well managed ex situ collections. Ex situ conservation focuses on safeguarding species by keeping them in places such as seed banks or living collections. In situ means "on site", so in situ conservation is the conservation of species diversity within normal and natural habitats and ecosystems. The Scott Arboretum is a member of Botanical Gardens Conservation International (BGCI), which works with botanic gardens around the world and other conservation partners to secure plant diversity for the benefit of people and the planet. The aim of BGCI is to ensure that threatened species are secure in botanic garden collections as an insurance policy against loss in the wild. Their work encompasses supporting botanic garden development where this is needed and addressing capacity building needs. They support ex situ conservation for priority species, with a focus on linking ex situ conservation with species conservation in natural habitats and they work with botanic gardens on the development and implementation of habitat restoration and education projects. BGCI uses the IUCN Red List of Threatened Species™ to identify the level of threat to plants. In-depth analyses of the data contained in the IUCN, the International Union for Conservation of Nature, Red List are published periodically (usually at least once every four years). The results from the analysis of the data contained in the 2008 update of the IUCN Red List are published in The 2008 Review of the IUCN Red List of Threatened Species; see www.iucn.org/redlist for further details. -

Amur Oblast TYNDINSKY 361,900 Sq
AMUR 196 Ⅲ THE RUSSIAN FAR EAST SAKHA Map 5.1 Ust-Nyukzha Amur Oblast TY NDINS KY 361,900 sq. km Lopcha Lapri Ust-Urkima Baikal-Amur Mainline Tynda CHITA !. ZEISKY Kirovsky Kirovsky Zeiskoe Zolotaya Gora Reservoir Takhtamygda Solovyovsk Urkan Urusha !Skovorodino KHABAROVSK Erofei Pavlovich Never SKOVO MAGDAGACHINSKY Tra ns-Siberian Railroad DIRO Taldan Mokhe NSKY Zeya .! Ignashino Ivanovka Dzhalinda Ovsyanka ! Pioner Magdagachi Beketovo Yasny Tolbuzino Yubileiny Tokur Ekimchan Tygda Inzhan Oktyabrskiy Lukachek Zlatoustovsk Koboldo Ushumun Stoiba Ivanovskoe Chernyaevo Sivaki Ogodzha Ust-Tygda Selemdzhinsk Kuznetsovo Byssa Fevralsk KY Kukhterin-Lug NS Mukhino Tu Novorossiika Norsk M DHI Chagoyan Maisky SELE Novovoskresenovka SKY N OV ! Shimanovsk Uglovoe MAZ SHIMA ANOV Novogeorgievka Y Novokievsky Uval SK EN SK Mazanovo Y SVOBODN Chernigovka !. Svobodny Margaritovka e CHINA Kostyukovka inlin SERYSHEVSKY ! Seryshevo Belogorsk ROMNENSKY rMa Bolshaya Sazanka !. Shiroky Log - Amu BELOGORSKY Pridorozhnoe BLAGOVESHCHENSKY Romny Baikal Pozdeevka Berezovka Novotroitskoe IVANOVSKY Ekaterinoslavka Y Cheugda Ivanovka Talakan BRSKY SKY P! O KTYA INSK EI BLAGOVESHCHENSK Tambovka ZavitinskIT BUR ! Bakhirevo ZAV T A M B OVSKY Muravyovka Raichikhinsk ! ! VKONSTANTINO SKY Poyarkovo Progress ARKHARINSKY Konstantinovka Arkhara ! Gribovka M LIKHAI O VSKY ¯ Kundur Innokentevka Leninskoe km A m Trans -Siberianad Railro u 100 r R i v JAO Russian Far East e r By Newell and Zhou / Sources: Ministry of Natural Resources, 2002; ESRI, 2002. Newell, J. 2004. The Russian Far East: A Reference Guide for Conservation and Development. McKinleyville, CA: Daniel & Daniel. 466 pages CHAPTER 5 Amur Oblast Location Amur Oblast, in the upper and middle Amur River basin, is 8,000 km east of Moscow by rail (or 6,500 km by air). -

Formation of Spatial Mosaic of Abies Nephrolepis (Pinaceae) Populations in Korean Pine- Broadleaved Forests in the South of Russian Far East © T
Rastitelnye Resursy. 53(4): 480—495, 2017 FORMATION OF SPATIAL MOSAIC OF ABIES NEPHROLEPIS (PINACEAE) POPULATIONS IN KOREAN PINE- BROADLEAVED FORESTS IN THE SOUTH OF RUSSIAN FAR EAST © T. Ya. Petrenko, *, 1, 2 A. M. Omelko, 1A. A. Zhmerenetsky, 1 O. N. Ukhvatkina,1 L. A. Sibirina1 1 Federal Scientific center of the East Asia terrestrial biodiversity FEB RAS, Vladivostok, Russia 2Far Eastern Federal University, Vladivostok, Russia *E-mail: [email protected] REFERENCES 1. Ricklefs R. E. 1990. Ecology. Heidelberg. 896 p. 2. Hubbel S. P. 2001. The unified neutral theory of biodiversity and biogeography. Princeton. 390 p. 3. Whittaker R. H., Levin S. A. 1977. The role of mosaic phenomena in natural communities. –– Theoret. Pop. Biol. 12: 117–139. 4. Hao Z., Zhang J., Song B., Ye J., Li B. 2007.Vertical structure and spatial associations of dominant tree species in an old-growth temperate forest. –– For. Ecol. Manag. 252: 1–11. 5. Krestov P. V. 2003. Forest vegetation for Easternmost Russia (Russian Far East). –– For. Veg. Northeast Asia. Dordrecht. P. 93–180. 6. Liu Y. Y., Jin G. Z. 2012. Spatial distribution patterns and dynamics of four dominant tree species in a typical mixed broadleaved-Korean pine forest. –– J. Pl. Interact. 9(1): 745–753. 7. Kolesnikov B. P. 1956. Kedrovyye lesa Dalnego Vostoka [Korean pine forest of the Far East]. Moscow. 262 p. (In Russian) 8. Usenko N. V. 1984. Derevya, kustarniki i liany DalnegoVostoka [Trees, shrubs and lianas of the Far East]. Khabarovsk. 272 p. (In Russian) 9. Ishikawa Y., Krestov P. V., Namikawa K. 1999. -

Evolution of the Female Conifer Cone Fossils, Morphology and Phylogenetics
DEPARTMENT OF BIOLOGICAL AND ENVIRONMENTAL SCIENCES EVOLUTION OF THE FEMALE CONIFER CONE FOSSILS, MORPHOLOGY AND PHYLOGENETICS Daniel Bäck Degree project for Bachelor of Science with a major in Biology BIO602, Biologi: Examensarbete – kandidatexamen, 15 hp First cycle Semester/year: Spring 2020 Supervisor: Åslög Dahl, Department of Biological and Environmental Sciences Examiner: Claes Persson, Department of Biological and Environmental Sciences Front page: Abies koreana (immature seed cones), Gothenburg Botanical Garden, Sweden Table of contents 1 Abstract ............................................................................................................................... 2 2 Introduction ......................................................................................................................... 3 2.1 Brief history of Florin’s research ............................................................................... 3 2.2 Progress in conifer phylogenetics .............................................................................. 4 3 Aims .................................................................................................................................... 4 4 Materials and Methods ........................................................................................................ 4 4.1 Literature: ................................................................................................................... 4 4.2 RStudio: ..................................................................................................................... -
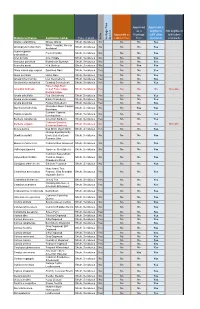
Botanical Name Common Name
Approved Approved & as a eligible to Not eligible to Approved as Frontage fulfill other fulfill other Type of plant a Street Tree Tree standards standards Heritage Tree Tree Heritage Species Botanical Name Common name Native Abelia x grandiflora Glossy Abelia Shrub, Deciduous No No No Yes White Forsytha; Korean Abeliophyllum distichum Shrub, Deciduous No No No Yes Abelialeaf Acanthropanax Fiveleaf Aralia Shrub, Deciduous No No No Yes sieboldianus Acer ginnala Amur Maple Shrub, Deciduous No No No Yes Aesculus parviflora Bottlebrush Buckeye Shrub, Deciduous No No No Yes Aesculus pavia Red Buckeye Shrub, Deciduous No No Yes Yes Alnus incana ssp. rugosa Speckled Alder Shrub, Deciduous Yes No No Yes Alnus serrulata Hazel Alder Shrub, Deciduous Yes No No Yes Amelanchier humilis Low Serviceberry Shrub, Deciduous Yes No No Yes Amelanchier stolonifera Running Serviceberry Shrub, Deciduous Yes No No Yes False Indigo Bush; Amorpha fruticosa Desert False Indigo; Shrub, Deciduous Yes No No No Not eligible Bastard Indigo Aronia arbutifolia Red Chokeberry Shrub, Deciduous Yes No No Yes Aronia melanocarpa Black Chokeberry Shrub, Deciduous Yes No No Yes Aronia prunifolia Purple Chokeberry Shrub, Deciduous Yes No No Yes Groundsel-Bush; Eastern Baccharis halimifolia Shrub, Deciduous No No Yes Yes Baccharis Summer Cypress; Bassia scoparia Shrub, Deciduous No No No Yes Burning-Bush Berberis canadensis American Barberry Shrub, Deciduous Yes No No Yes Common Barberry; Berberis vulgaris Shrub, Deciduous No No No No Not eligible European Barberry Betula pumila -

Genetic Variation in Natural Populations of Abies Nephrolepis Max
Genetic variation in natural populations of Abies nephrolepis Max. in South Korea Lee Seok Woo, Yang Byeong Hoon, Han Sang Don, Song Jung Ho, Lee Jung Joo To cite this version: Lee Seok Woo, Yang Byeong Hoon, Han Sang Don, Song Jung Ho, Lee Jung Joo. Genetic variation in natural populations of Abies nephrolepis Max. in South Korea. Annals of Forest Science, Springer Nature (since 2011)/EDP Science (until 2010), 2008, 65 (3), pp.1. hal-00883380 HAL Id: hal-00883380 https://hal.archives-ouvertes.fr/hal-00883380 Submitted on 1 Jan 2008 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Ann. For. Sci. 65 (2008) 302 Available online at: c INRA, EDP Sciences, 2008 www.afs-journal.org DOI: 10.1051/forest:2008006 Original article Genetic variation in natural populations of Abies nephrolepis Max. in South Korea Lee Seok Woo*, Yang Byeong Hoon,HanSangDon, Song Jung Ho, Lee Jung Joo Department of Forest Genetic Resources, Korea Forest Research Institute, Suwon 441-350, Rep. of Korea (Received 22 June 2007; accepted 12 November 2007) Abstract – Abies nephrolepis Max. is a fir species occurring in Northeast China, the extreme southeast of Russia and Korea. -

Diplopoda) from the Asian Part of Russia
Number 316: 1-25 ISSN 1026-051X August 2016 http/urn:lsid:zoobank.org:pub:2F078599-97CF-4A0E-8E89-285F43A5B5F5 NEW SPECIES AND NEW RECORDS OF MILLIPEDES (DIPLOPODA) FROM THE ASIAN PART OF RUSSIA E. V. Mikhaljova Institute of Biology and Soil Science, Far Eastern Branch of the Russian Academy of Sciences, Vladivostok 690022, Russia. E-mail: [email protected] Pacifiosoma asperum Mikhaljova, sp. n. and Diplomaragna konyukhovi Mikhaljova, sp. n. are described from the Russian Far East (Khabarovskii krai and Primorskii krai). New faunistic records for the Asian part of Russia are given for other millipede species. The record of East Asian Epanerchodus orientalis Attems, 1901 in Kunashir Island (Gongalsky et al., 2014) is based on misidentification and belongs to Epanerchodus kunashiricus Mikhaljova, 1988. KEY WORDS: Diplopoda, taxonomy, fauna, new species, new records, Siberia, Russian Far East. Е. В. Михалёва. Новые виды и новые фаунистические находки дву- парноногих многоножек (Diplopoda) азиатской части России // Дальне- восточный энтомолог. 2016. N 316. С. 1-25. Из Хабаровского и Приморского краев описаны Pacifiosoma asperum Mikhaljova, sp. n. и Diplomaragna konyukhovi Mikhaljova, sp. n. Приведены новые фаунистические находки других видов двупарноногих многоножек. Указание восточноазиатского Epanerchodus orientalis Attems, 1901 для острова Кунашир (Гонгальский и др., 2014) ошибочно и относится к Epanerchodus kunashiricus Mikhaljova, 1988. Биолого-почвенный институт ДВО РАН, Владивосток, 690022, Россия. 1 INTRODUCTION This contribution continues researches in the millipede fauna of the huge areas of the Asian part of Russia, this time being confined to the abundant material accumulated during the last decade. The paper is devoted to new records of the known, as well as to descriptions of two new species and to the correction of an error of species identification. -

Genetic Variation in Natural Populations of Abies Nephrolepis Max
Ann. For. Sci. 65 (2008) 302 Available online at: c INRA, EDP Sciences, 2008 www.afs-journal.org DOI: 10.1051/forest:2008006 Original article Genetic variation in natural populations of Abies nephrolepis Max. in South Korea Lee Seok Woo*, Yang Byeong Hoon,HanSangDon, Song Jung Ho, Lee Jung Joo Department of Forest Genetic Resources, Korea Forest Research Institute, Suwon 441-350, Rep. of Korea (Received 22 June 2007; accepted 12 November 2007) Abstract – Abies nephrolepis Max. is a fir species occurring in Northeast China, the extreme southeast of Russia and Korea. In Korea, A. nephrolepis is one of only three native fir species growing in high, mountainous areas with a disjunct distribution. We investigated genetic variation in A. nephrolepis by examining 24 ISSR polymorphic loci in 248 individuals representing eight natural populations in Korea. The level of intra-population genetic diversity (He = 0.240) was similar to or slightly lower than that of plants with similar ecological traits and life history. Most of the genetic diversity was allocated among individuals within populations (GST = 0.082, ΦST = 0.041) and the number of migrants per generation (Nm) was 5.56. Nei’s genetic distances were small (D = 0.027) and unrelated to geographic distances between populations. Implications for the conservation of genetic variation in A. nephrolepis in South Korea were discussed. Abies nephrolepis Max. / ISSR / genetic diversity / genetic differentiation Résumé – Variabilité génétique dans des populations naturelles de Abies nephrolepis Max. en Coréee du Sud. Abies nephrolepis Max. est un sapin présent dans le Nord-Est de la Chine, l’extrême Sud-Est de la Russie et en Corée. -

The Evolution of Cavitation Resistance in Conifers Maximilian Larter
The evolution of cavitation resistance in conifers Maximilian Larter To cite this version: Maximilian Larter. The evolution of cavitation resistance in conifers. Bioclimatology. Univer- sit´ede Bordeaux, 2016. English. <NNT : 2016BORD0103>. <tel-01375936> HAL Id: tel-01375936 https://tel.archives-ouvertes.fr/tel-01375936 Submitted on 3 Oct 2016 HAL is a multi-disciplinary open access L'archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destin´eeau d´ep^otet `ala diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publi´esou non, lished or not. The documents may come from ´emanant des ´etablissements d'enseignement et de teaching and research institutions in France or recherche fran¸caisou ´etrangers,des laboratoires abroad, or from public or private research centers. publics ou priv´es. THESE Pour obtenir le grade de DOCTEUR DE L’UNIVERSITE DE BORDEAUX Spécialité : Ecologie évolutive, fonctionnelle et des communautés Ecole doctorale: Sciences et Environnements Evolution de la résistance à la cavitation chez les conifères The evolution of cavitation resistance in conifers Maximilian LARTER Directeur : Sylvain DELZON (DR INRA) Co-Directeur : Jean-Christophe DOMEC (Professeur, BSA) Soutenue le 22/07/2016 Devant le jury composé de : Rapporteurs : Mme Amy ZANNE, Prof., George Washington University Mr Jordi MARTINEZ VILALTA, Prof., Universitat Autonoma de Barcelona Examinateurs : Mme Lisa WINGATE, CR INRA, UMR ISPA, Bordeaux Mr Jérôme CHAVE, DR CNRS, UMR EDB, Toulouse i ii Abstract Title: The evolution of cavitation resistance in conifers Abstract Forests worldwide are at increased risk of widespread mortality due to intense drought under current and future climate change.