The Complete Chloroplast Genome Sequence of Abies Nephrolepis (Pinaceae: Abietoideae)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Programa Nacional Para La Aplicación De La Normativa Fitosanitaria
PROGRAMA NACIONAL PARA LA APLICACIÓN DE LA NORMATIVA FITOSANITARIA PLAN NACIONAL DE CONTINGENCIA DE Dendrolimus sibiricus Tschetverikov SEPTIEMBRE 2020 SUMARIO DE MODIFICACIONES REVISIÓN FECHA DESCRIPCIÓN OBJETO DE LA REVISIÓN 30/09/2020 Documento base Plan Contingencia Dendrolimus sibiricus 2020 INDICE 1. Introducción y Objetivos 2. Definiciones 3. Marco legislativo, Organización y Estructura de mando 3.1 Marco legislativo 3.2 Marco legislativo, Organización y Estructura 4. Información sobre la enfermedad 4.1 Distribución de la plaga 4.2 Taxonomía 4.3 Daño 4.4 Plantas hospedantes 5. Métodos de identificación y diagnóstico 5.1 Detección de la plaga 5.2 Identificación y diagnóstico 6. Ejecución del Plan Nacional de Contingencia 6.1 Plan de Nacional Contingencia y los planes específicos de acción 6.2 Medidas cautelares a adoptar en caso de sospecha de la presencia de Dendrolimus sibiricus. 6.3 Medidas a adoptar en caso de confirmación de la presencia de Dendrolimus sibiricus. 6.4 Medidas de erradicación. 6.5 Medidas en caso de incumplimiento. 7. Comunicación, Documentación y Formación 7.1. Comunicación externa y campañas de divulgación/sensibilización. 7.2 Consulta a los grupos de interés 7.3. Comunicación interna y documentación 7.4. Pruebas y formación del personal 8. Evaluación y revisión 9. Referencias Anexo 1: PROTOCOLO DE PROSPECCIONES DE Dendrolimus sibiricus Anexo 2: PROGRAMA DE ERRADICACIÓN DE Dendrolimus sibiricus Página 1 de 27 Plan Contingencia Dendrolimus sibiricus 2020 1. Introducción y Objetivos En el presente documento se recogen las medidas que deben adoptarse contra Dendrolimus sibiricus, organismo nocivo regulado, con el objetivo de impedir su aparición, y en caso de que aparezca, actuar con rapidez y eficacia, determinar su distribución y aplicar medidas de erradicación. -

Potential Impact of Climate Change
Adhikari et al. Journal of Ecology and Environment (2018) 42:36 Journal of Ecology https://doi.org/10.1186/s41610-018-0095-y and Environment RESEARCH Open Access Potential impact of climate change on the species richness of subalpine plant species in the mountain national parks of South Korea Pradeep Adhikari, Man-Seok Shin, Ja-Young Jeon, Hyun Woo Kim, Seungbum Hong and Changwan Seo* Abstract Background: Subalpine ecosystems at high altitudes and latitudes are particularly sensitive to climate change. In South Korea, the prediction of the species richness of subalpine plant species under future climate change is not well studied. Thus, this study aims to assess the potential impact of climate change on species richness of subalpine plant species (14 species) in the 17 mountain national parks (MNPs) of South Korea under climate change scenarios’ representative concentration pathways (RCP) 4.5 and RCP 8.5 using maximum entropy (MaxEnt) and Migclim for the years 2050 and 2070. Results: Altogether, 723 species occurrence points of 14 species and six selected variables were used in modeling. The models developed for all species showed excellent performance (AUC > 0.89 and TSS > 0.70). The results predicted a significant loss of species richness in all MNPs. Under RCP 4.5, the range of reduction was predicted to be 15.38–94.02% by 2050 and 21.42–96.64% by 2070. Similarly, under RCP 8.5, it will decline 15.38–97.9% by 2050 and 23.07–100% by 2070. The reduction was relatively high in the MNPs located in the central regions (Songnisan and Gyeryongsan), eastern region (Juwangsan), and southern regions (Mudeungsan, Wolchulsan, Hallasan, and Jirisan) compared to the northern and northeastern regions (Odaesan, Seoraksan, Chiaksan, and Taebaeksan). -

Stand Structure and Dynamics During a 16-Year Period in a Conifer-Hardwood Mixed Forest, Northern Japan
Takahashi et al. 1 Stand structure and dynamics during a 16-year period in a sub-boreal conifer-hardwood mixed forest, northern Japan Koichi Takahashia,1,*, Daisuke Mitsuishia, Shigeru Uemurab, Jun-Ichirou Suzukia,2, Toshihiko Haraa a: The Institute of Low Temperature Science, Hokkaido University, Sapporo 060-0819, Japan b: Forest Research Station, Field Science Center for Northern Biosphere, Hokkaido University, Nayoro 096-0071, Japan Present address 1: Department of Biology, Faculty of Science, Shinshu University, Matsumoto 390-8621, Japan 2: Department of Biology, Faculty of Science, Tokyo Metropolitan University, Tokyo 192-0397, Japan *: All correspondence to K. Takahashi at the above present address#1. Fax: +81.263.37.2560 E-mail: [email protected] This manuscript consists of 23-typed pages including figure legends, 3 tables and 6 figures. Takahashi et al. 2 Abstract The stand structure and regeneration dynamics of trees > 2.0 m in trunk height were studied during 1982–1998 in a 1-ha plot in a sub-boreal conifer-hardwood mixed forest, northern Japan, with a dense dwarf bamboo in the understory. Total density was low in 1982 (651 trees/ha), as compared with other forests in Japan. Quercus crispula was dominant in the upper canopy layer but their saplings were rare, while Acer mono, Acer japonicum and Abies sachalinensis were dominant in the sub-canopy and understory layers with many saplings. Mortality of each species was quite low during the census period (average 0.57%/yr), and there was no clear difference among the four species. The age structure of Quercus crispula was bell-shaped with a peak at ca. -

Phylogeny and Biogeography of Tsuga (Pinaceae)
Systematic Botany (2008), 33(3): pp. 478–489 © Copyright 2008 by the American Society of Plant Taxonomists Phylogeny and Biogeography of Tsuga (Pinaceae) Inferred from Nuclear Ribosomal ITS and Chloroplast DNA Sequence Data Nathan P. Havill1,6, Christopher S. Campbell2, Thomas F. Vining2,5, Ben LePage3, Randall J. Bayer4, and Michael J. Donoghue1 1Department of Ecology and Evolutionary Biology, Yale University, New Haven, Connecticut 06520-8106 U.S.A 2School of Biology and Ecology, University of Maine, Orono, Maine 04469-5735 U.S.A. 3The Academy of Natural Sciences, 1900 Benjamin Franklin Parkway, Philadelphia, Pennsylvania 19103 U.S.A. 4CSIRO – Division of Plant Industry, Center for Plant Biodiversity Research, GPO 1600, Canberra, ACT 2601 Australia; present address: Department of Biology, University of Memphis, Memphis, Tennesee 38152 U.S.A. 5Present address: Delta Institute of Natural History, 219 Dead River Road, Bowdoin, Maine 04287 U.S.A. 6Author for correspondence ([email protected]) Communicating Editor: Matt Lavin Abstract—Hemlock, Tsuga (Pinaceae), has a disjunct distribution in North America and Asia. To examine the biogeographic history of Tsuga, phylogenetic relationships among multiple accessions of all nine species were inferred using chloroplast DNA sequences and multiple cloned sequences of the nuclear ribosomal ITS region. Analysis of chloroplast and ITS sequences resolve a clade that includes the two western North American species, T. heterophylla and T. mertensiana, and a clade of Asian species within which one of the eastern North American species, T. caroliniana, is nested. The other eastern North American species, T. canadensis, is sister to the Asian clade. Tsuga chinensis from Taiwan did not group with T. -

Ten Years of Provenance Trials and Application of Multivariate Random Forests Predicted the Most Preferable Seed Source for Silv
Article Ten Years of Provenance Trials and Application of Multivariate Random Forests Predicted the Most Preferable Seed Source for Silviculture of Abies sachalinensis in Hokkaido, Japan Ikutaro Tsuyama 1,*, Wataru Ishizuka 2 , Keiko Kitamura 1, Haruhiko Taneda 3 and Susumu Goto 4 1 Hokkaido Research Center, Forestry and Forest Products Research Institute, 7 Hitsujigaoka, Toyohira, Sapporo, Hokkaido 062-8516, Japan; [email protected] 2 Forestry Research Institute, Hokkaido Research Organization, Koushunai, Bibai, Hokkaido 079-0198, Japan; [email protected] 3 Department of Biological Sciences, Graduate School of Science, The University of Tokyo, 7-3-1, Hongo, Bunkyo, Tokyo 113-0033, Japan; [email protected] 4 Education and Research Center, The University of Tokyo Forests, Graduate School of Agricultural and Life Sciences, The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan; [email protected] * Correspondence: [email protected] Received: 10 August 2020; Accepted: 27 September 2020; Published: 30 September 2020 Abstract: Research highlights: Using 10-year tree height data obtained after planting from the range-wide provenance trials of Abies sachalinensis, we constructed multivariate random forests (MRF), a machine learning algorithm, with climatic variables. The constructed MRF enabled prediction of the optimum seed source to achieve good performance in terms of height growth at every planting site on a fine scale. Background and objectives: Because forest tree species are adapted to the local environment, local seeds are empirically considered as the best sources for planting. However, in some cases, local seed sources show lower performance in height growth than that showed by non-local seed sources. -

ISTA List of Stabilized Plant Names 7Th Edition
ISTA List of Stabilized Plant Names th 7 Edition ISTA Nomenclature Committee Chair: Dr. M. Schori Published by All rights reserved. No part of this publication may be The Internation Seed Testing Association (ISTA) reproduced, stored in any retrieval system or transmitted Zürichstr. 50, CH-8303 Bassersdorf, Switzerland in any form or by any means, electronic, mechanical, photocopying, recording or otherwise, without prior ©2020 International Seed Testing Association (ISTA) permission in writing from ISTA. ISBN 978-3-906549-77-4 ISTA List of Stabilized Plant Names 1st Edition 1966 ISTA Nomenclature Committee Chair: Prof P. A. Linehan 2nd Edition 1983 ISTA Nomenclature Committee Chair: Dr. H. Pirson 3rd Edition 1988 ISTA Nomenclature Committee Chair: Dr. W. A. Brandenburg 4th Edition 2001 ISTA Nomenclature Committee Chair: Dr. J. H. Wiersema 5th Edition 2007 ISTA Nomenclature Committee Chair: Dr. J. H. Wiersema 6th Edition 2013 ISTA Nomenclature Committee Chair: Dr. J. H. Wiersema 7th Edition 2019 ISTA Nomenclature Committee Chair: Dr. M. Schori 2 7th Edition ISTA List of Stabilized Plant Names Content Preface .......................................................................................................................................................... 4 Acknowledgements ....................................................................................................................................... 6 Symbols and Abbreviations .......................................................................................................................... -
IUCN Red List of Threatened Species™ to Identify the Level of Threat to Plants
Ex-Situ Conservation at Scott Arboretum Public gardens and arboreta are more than just pretty places. They serve as an insurance policy for the future through their well managed ex situ collections. Ex situ conservation focuses on safeguarding species by keeping them in places such as seed banks or living collections. In situ means "on site", so in situ conservation is the conservation of species diversity within normal and natural habitats and ecosystems. The Scott Arboretum is a member of Botanical Gardens Conservation International (BGCI), which works with botanic gardens around the world and other conservation partners to secure plant diversity for the benefit of people and the planet. The aim of BGCI is to ensure that threatened species are secure in botanic garden collections as an insurance policy against loss in the wild. Their work encompasses supporting botanic garden development where this is needed and addressing capacity building needs. They support ex situ conservation for priority species, with a focus on linking ex situ conservation with species conservation in natural habitats and they work with botanic gardens on the development and implementation of habitat restoration and education projects. BGCI uses the IUCN Red List of Threatened Species™ to identify the level of threat to plants. In-depth analyses of the data contained in the IUCN, the International Union for Conservation of Nature, Red List are published periodically (usually at least once every four years). The results from the analysis of the data contained in the 2008 update of the IUCN Red List are published in The 2008 Review of the IUCN Red List of Threatened Species; see www.iucn.org/redlist for further details. -

Geographical Gradients of Genetic Diversity and Differentiation Among the Southernmost Marginal Populations of Abies Sachalinens
Article Geographical Gradients of Genetic Diversity and Differentiation among the Southernmost Marginal Populations of Abies sachalinensis Revealed by EST-SSR Polymorphism Keiko Kitamura 1, Kentaro Uchiyama 2, Saneyoshi Ueno 2, Wataru Ishizuka 3, Ikutaro Tsuyama 1 and Susumu Goto 4,* 1 Hokkaido Research Center, Forestry and Forest Products Research Institute, 7 Hitsujigaoka, Toyohira, Sapporo, Hokkaido 062-8516, Japan; kitamq@ffpri.affrc.go.jp (K.K.); itsuyama@affrc.go.jp (T.I.) 2 Department of Forest Molecular Genetics and Biotechnology, Forestry and Forest Products Research Institute, 1 Matsunosato, Tsukuba, Ibaraki 305-8687, Japan; [email protected] (U.K.); [email protected] (U.S.) 3 Forestry Research Institute, Hokkaido Research Organization, Koushunai, Bibai, Hokkaido 079-0166, Japan; [email protected] 4 Education and Research Center, The University of Tokyo Forests, Graduate School of Agricultural and Life Sciences, The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan * Correspondence: [email protected] Received: 17 December 2019; Accepted: 17 February 2020; Published: 20 February 2020 Abstract: Research Highlights: We detected the longitudinal gradients of genetic diversity parameters, such as the number of alleles, effective number of alleles, heterozygosity, and inbreeding coefficient, and found that these might be attributable to climatic conditions, such as temperature and snow depth. Background and Objectives: Genetic diversity among local populations of a plant species at its distributional margin has long been of interest in ecological genetics. Populations at the distribution center grow well in favorable conditions, but those at the range margins are exposed to unfavorable environments, and the environmental conditions at establishment sites might reflect the genetic diversity of local populations. -

Evolution of the Female Conifer Cone Fossils, Morphology and Phylogenetics
DEPARTMENT OF BIOLOGICAL AND ENVIRONMENTAL SCIENCES EVOLUTION OF THE FEMALE CONIFER CONE FOSSILS, MORPHOLOGY AND PHYLOGENETICS Daniel Bäck Degree project for Bachelor of Science with a major in Biology BIO602, Biologi: Examensarbete – kandidatexamen, 15 hp First cycle Semester/year: Spring 2020 Supervisor: Åslög Dahl, Department of Biological and Environmental Sciences Examiner: Claes Persson, Department of Biological and Environmental Sciences Front page: Abies koreana (immature seed cones), Gothenburg Botanical Garden, Sweden Table of contents 1 Abstract ............................................................................................................................... 2 2 Introduction ......................................................................................................................... 3 2.1 Brief history of Florin’s research ............................................................................... 3 2.2 Progress in conifer phylogenetics .............................................................................. 4 3 Aims .................................................................................................................................... 4 4 Materials and Methods ........................................................................................................ 4 4.1 Literature: ................................................................................................................... 4 4.2 RStudio: ..................................................................................................................... -
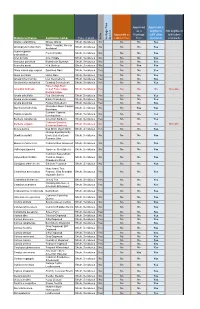
Botanical Name Common Name
Approved Approved & as a eligible to Not eligible to Approved as Frontage fulfill other fulfill other Type of plant a Street Tree Tree standards standards Heritage Tree Tree Heritage Species Botanical Name Common name Native Abelia x grandiflora Glossy Abelia Shrub, Deciduous No No No Yes White Forsytha; Korean Abeliophyllum distichum Shrub, Deciduous No No No Yes Abelialeaf Acanthropanax Fiveleaf Aralia Shrub, Deciduous No No No Yes sieboldianus Acer ginnala Amur Maple Shrub, Deciduous No No No Yes Aesculus parviflora Bottlebrush Buckeye Shrub, Deciduous No No No Yes Aesculus pavia Red Buckeye Shrub, Deciduous No No Yes Yes Alnus incana ssp. rugosa Speckled Alder Shrub, Deciduous Yes No No Yes Alnus serrulata Hazel Alder Shrub, Deciduous Yes No No Yes Amelanchier humilis Low Serviceberry Shrub, Deciduous Yes No No Yes Amelanchier stolonifera Running Serviceberry Shrub, Deciduous Yes No No Yes False Indigo Bush; Amorpha fruticosa Desert False Indigo; Shrub, Deciduous Yes No No No Not eligible Bastard Indigo Aronia arbutifolia Red Chokeberry Shrub, Deciduous Yes No No Yes Aronia melanocarpa Black Chokeberry Shrub, Deciduous Yes No No Yes Aronia prunifolia Purple Chokeberry Shrub, Deciduous Yes No No Yes Groundsel-Bush; Eastern Baccharis halimifolia Shrub, Deciduous No No Yes Yes Baccharis Summer Cypress; Bassia scoparia Shrub, Deciduous No No No Yes Burning-Bush Berberis canadensis American Barberry Shrub, Deciduous Yes No No Yes Common Barberry; Berberis vulgaris Shrub, Deciduous No No No No Not eligible European Barberry Betula pumila -

Magnolia Obovata
ISSUE 80 INAGNOLN INagnolla obovata Eric Hsu, Putnam Fellow, Arnold Arboretum of Harvard University Photographs by Philippe de 8 poelberch I first encountered Magnolia obovata in Bower at Sir Harold Hillier Gardens and Arboretum, Hampshire, England, where the tightly pursed, waxy, globular buds teased, but rewarded my patience. As each bud unfurled successively, it emitted an intoxicating ambrosial bouquet of melons, bananas, and grapes. Although the leaves were nowhere as luxuriously lustrous as M. grandrflora, they formed an el- egant wreath for the creamy white flower. I gingerly plucked one flower for doser observation, and placed one in my room. When I re- tumed from work later in the afternoon, the mom was overpowering- ly redolent of the magnolia's scent. The same olfactory pleasure was later experienced vicariously through the large Magnolia x wiesneri in the private garden of Nicholas Nickou in southern Connecticut. Several years earlier, I had traveled to Hokkaido Japan, after my high school graduation. Although Hokkaido experiences more severe win- ters than those in the southern parts of Japan, the forests there yield a remarkable diversity of fora, some of which are popular ornamen- tals. When one drives through the region, the silvery to blue-green leaf undersides of Magnolia obovata, shimmering in the breeze, seem to flag the eyes. In "Forest Flora of Japan" (sggII), Charles Sargent commended this species, which he encountered growing tluough the mountainous forests of Hokkaido. He called it "one of the largest and most beautiful of the deciduous-leaved species in size and [the seed conesj are sometimes eight inches long, and brilliant scarlet in color, stand out on branches, it is the most striking feature of the forests. -

Ectomycorrhization of Tricholoma Matsutake with Abies Veitchii and Tsuga Diversifolia in the Subalpine Forests of Japan
mycoscience 56 (2015) 402e412 Available online at www.sciencedirect.com journal homepage: www.elsevier.com/locate/myc Full paper Ectomycorrhization of Tricholoma matsutake with Abies veitchii and Tsuga diversifolia in the subalpine forests of Japan Naoki Endo a, Preeyaporn Dokmai b,c, Nuttika Suwannasai b, Cherdchai Phosri d, Yuka Horimai c, Nobuhiro Hirai e, Masaki Fukuda a,c, * Akiyoshi Yamada a,c,f, a Department of Bioscience and Food Production Science, Interdisciplinary Graduate School of Science and Technology, Shinshu University, 8304, Minami-minowa, Nagano, 399-4598, Japan b Faculty of Science, Srinakharinwirot University, 114, Sukhumvit 23, Bangkok, 10110, Thailand c Department of Bioscience and Biotechnology, Faculty of Agriculture, Shinshu University, 8304, Minami-minowa, Nagano, 399-4598, Japan d Division of Science, Faculty of Liberal Arts and Science, Nakhon Phanom University, 167 Naratchakouy Sub- District, Muang District, Nakhon Phanom, 48000, Thailand e Graduate School of Agriculture, Kyoto University, Kitashirakawa-Oiwake-cho, Sakyo-ku, Kyoto, 606-8502, Japan f Division of Terrestrial Ecosystem, Institute of Mountain Science, Shinshu University, 8304, Minami-minowa, Nagano, 399-4598, Japan article info abstract Article history: Tricholoma matsutake is one of the most highly valued ectomycorrhizal mushrooms, pri- Received 15 July 2014 marily associated with conifers. Here, we examined the association of T. matsutake with Received in revised form hemlock and fir species native to the subalpine forests of Japan. Basidiomata of T. matsu- 12 December 2014 take were harvested from the forests of Honshu Island, Japan, along with two soil samples Accepted 15 December 2014 directly beneath the basidiomata; ectomycorrhizal root tips were also collected, and used Available online 28 January 2015 for fungal isolation and molecular analyses.