Fundulus Heteroclitus): a Phylogenetic and Population-Based Comparison
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplemental Information to Mammadova-Bach Et Al., “Laminin Α1 Orchestrates VEGFA Functions in the Ecosystem of Colorectal Carcinogenesis”
Supplemental information to Mammadova-Bach et al., “Laminin α1 orchestrates VEGFA functions in the ecosystem of colorectal carcinogenesis” Supplemental material and methods Cloning of the villin-LMα1 vector The plasmid pBS-villin-promoter containing the 3.5 Kb of the murine villin promoter, the first non coding exon, 5.5 kb of the first intron and 15 nucleotides of the second villin exon, was generated by S. Robine (Institut Curie, Paris, France). The EcoRI site in the multi cloning site was destroyed by fill in ligation with T4 polymerase according to the manufacturer`s instructions (New England Biolabs, Ozyme, Saint Quentin en Yvelines, France). Site directed mutagenesis (GeneEditor in vitro Site-Directed Mutagenesis system, Promega, Charbonnières-les-Bains, France) was then used to introduce a BsiWI site before the start codon of the villin coding sequence using the 5’ phosphorylated primer: 5’CCTTCTCCTCTAGGCTCGCGTACGATGACGTCGGACTTGCGG3’. A double strand annealed oligonucleotide, 5’GGCCGGACGCGTGAATTCGTCGACGC3’ and 5’GGCCGCGTCGACGAATTCACGC GTCC3’ containing restriction site for MluI, EcoRI and SalI were inserted in the NotI site (present in the multi cloning site), generating the plasmid pBS-villin-promoter-MES. The SV40 polyA region of the pEGFP plasmid (Clontech, Ozyme, Saint Quentin Yvelines, France) was amplified by PCR using primers 5’GGCGCCTCTAGATCATAATCAGCCATA3’ and 5’GGCGCCCTTAAGATACATTGATGAGTT3’ before subcloning into the pGEMTeasy vector (Promega, Charbonnières-les-Bains, France). After EcoRI digestion, the SV40 polyA fragment was purified with the NucleoSpin Extract II kit (Machery-Nagel, Hoerdt, France) and then subcloned into the EcoRI site of the plasmid pBS-villin-promoter-MES. Site directed mutagenesis was used to introduce a BsiWI site (5’ phosphorylated AGCGCAGGGAGCGGCGGCCGTACGATGCGCGGCAGCGGCACG3’) before the initiation codon and a MluI site (5’ phosphorylated 1 CCCGGGCCTGAGCCCTAAACGCGTGCCAGCCTCTGCCCTTGG3’) after the stop codon in the full length cDNA coding for the mouse LMα1 in the pCIS vector (kindly provided by P. -

Ryanodine Binding to Ryr1. Final Report Edward M
Evaluation of GSK2487213A on [3H]Ryanodine Binding to RyR1. Final Report Edward M. Balog, PhD School of Applied Physiology Georgia Institute of Technology December 17, 2010 1 Summary The effects of GSK2487213A (213A) on RyR1 function was assessed via skeletal muscle heavy sarcoplasmic reticulum (HSR) [3H]ryanodine binding. 213A did not affect HSR ryanodine binding when Ca2+ was the sole RyR1 regulator. Stripping HSR of FKBP12.0, via treatment with FK506, increased Ca2+-activated ryanodine binding. Exogenous FKBP12.0 alone or in combination with 213A and or reducing agents failed to reverse the effects prior exposure to FK506. Including FK506 in the HSR [3H]ryanodine binding buffer dose-dependently increased ryanodine binding. 213A did not alter the FK506 dose response. Treating HSR with the nitric oxide donor, NOR-3, did not substantially reduce the FKBP12.0 content of the HSR. Prior exposure to NOR-3 enhanced Ca2+ stimulated HSR ryanodine binding. 213A reversed the NOR-3 activation of RyR1. These results suggest that 213A reverses nitrosylation-induced activation of RyR1 and the inhibition does not occur via stabilizing the interaction between RyR1 and FKBP12.0. Introduction Ryanodine receptors (RyR) are endo/sarcoplamic reticulum (SR) resident Ca2+ selective channels that form the efflux pathway for the release of Ca2+ from the SR to initiate muscle contraction. Isoform 1 (RyR1) is the predominate form in skeletal muscle, RyR2 is the predominate form in the heart while RyR3 has a widespread distribution. The channels are regulated by numerous endogenous effectors including ions primarily Ca2+ and Mg2+, metabolites such as the adenine nucleotides and accessory proteins including calmodulin and the FK506-binding proteins (FKBP). -

FKBP52 Regulates TRPC3-Dependent Ca<Sup>
© 2019. Published by The Company of Biologists Ltd | Journal of Cell Science (2019) 132, jcs231506. doi:10.1242/jcs.231506 RESEARCH ARTICLE FKBP52 regulates TRPC3-dependent Ca2+ signals and the hypertrophic growth of cardiomyocyte cultures Sandra Bandleon1, Patrick P. Strunz1, Simone Pickel2, Oleksandra Tiapko3, Antonella Cellini1, Erick Miranda-Laferte2 and Petra Eder-Negrin1,* ABSTRACT A single TRPC subunit is composed of six transmembrane The transient receptor potential (TRP; C-classical, TRPC) channel domains with a pore-forming loop connecting the transmembrane ‘ ’ TRPC3 allows a cation (Na+/Ca2+) influx that is favored by the domains 5 and 6, a preserved 25 amino acid sequence called a TRP domain and two cytosolic domains, an N-terminal ankyrin repeat stimulation of Gq protein-coupled receptors (GPCRs). An enhanced TRPC3 activity is related to adverse effects, including pathological domain and a C-terminal coiled-coil domain (Eder et al., 2007; Fan hypertrophy in chronic cardiac disease states. In the present study, et al., 2018). The cytosolic domains mediate ion channel formation we identified FK506-binding protein 52 (FKBP52, also known as and are implicated in ion channel regulation and plasma membrane FKBP4) as a novel interaction partner of TRPC3 in the heart. FKBP52 targeting (Eder et al., 2007). Among several protein interaction sites, was recovered from a cardiac cDNA library by a C-terminal TRPC3 the C-terminus of all TRPC subunits harbors a highly conserved fragment (amino acids 742–848) in a yeast two-hybrid screen. proline-rich sequence that corresponds to the binding domain in the Drosophila Downregulation of FKBP52 promoted a TRPC3-dependent photoreceptor channel TRPL for the FK506-binding hypertrophic response in neonatal rat cardiomyocytes (NRCs). -

TMEM95 Is a Sperm Membrane Protein Essential for Mammalian
RESEARCH ARTICLE TMEM95 is a sperm membrane protein essential for mammalian fertilization Ismael Lamas-Toranzo1†, Julieta G Hamze2†, Enrica Bianchi3, Beatriz Ferna´ ndez-Fuertes4,5, Serafı´nPe´ rez-Cerezales1, Ricardo Laguna-Barraza1, Rau´l Ferna´ ndez-Gonza´ lez1, Pat Lonergan4, Alfonso Gutie´ rrez-Ada´ n1, Gavin J Wright3, Marı´aJime´ nez-Movilla2*, Pablo Bermejo-A´ lvarez1* 1Animal Reproduction Department, INIA, Madrid, Spain; 2Department of Cell Biology and Histology, Medical School, University of Murcia, IMIB-Arrixaca, Murcia, Spain; 3Cell Surface Signalling Laboratory, Wellcome Trust Sanger Institute, Cambridge, United Kingdom; 4School of Agriculture and Food Science, University College Dublin, Dublin, Ireland; 5Department of Biology, Faculty of Sciences, Institute of Food and Agricultural Technology, University of Girona, Girona, Spain Abstract The fusion of gamete membranes during fertilization is an essential process for sexual reproduction. Despite its importance, only three proteins are known to be indispensable for sperm- egg membrane fusion: the sperm proteins IZUMO1 and SPACA6, and the egg protein JUNO. Here we demonstrate that another sperm protein, TMEM95, is necessary for sperm-egg interaction. TMEM95 ablation in mice caused complete male-specific infertility. Sperm lacking this protein were morphologically normal exhibited normal motility, and could penetrate the zona pellucida and bind to the oolemma. However, once bound to the oolemma, TMEM95-deficient sperm were unable to fuse with the egg membrane or penetrate into the ooplasm, and fertilization could only be achieved by mechanical injection of one sperm into the ooplasm, thereby bypassing membrane *For correspondence: fusion. These data demonstrate that TMEM95 is essential for mammalian fertilization. [email protected] (Mı´Je´-M); [email protected] (PB-A´ ) †These authors contributed equally to this work Introduction In sexually reproducing species, life begins with the fusion of two gametes during fertilization. -
Supplementary Tables and Figures
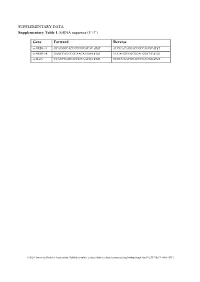
SUPPLEMENTARY DATA Supplementary Table 1. SiRNA sequence (5’-3’) Gene Forward Reverse si-HRD1-1# GCAUGGCAGUCCUGUACAU dTdT AUGUACAGGACUGCCAUGC dTdT si-HRD1-2# GAGCCAUCCGCAACAUGAA dTdT UUCAUGUUGCGGAUGGCUC dTdT si-MafA CCAUCGAGUACGUCAACGA dTdT UCGUUGACGUACUCGAUGG dTdT ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 2. Primer sequences for qRT-PCR (5’-3’) Gene Forward Reverse human HRD1 GCTCACGCCTACTACCTCAAA GCCAGACAAGTCTCTGTGACG mouse mafA AAGCGGCGCACGCTCAAGAA GGTCCCGCTCCTTGGCCAGA mouse insulin1 CACTTCCTACCCCTGCTGG ACCACAAAGATGCTGTTTGACA mouse β-actin AGGCCAACCGTGAAAAGATG AGAGCATAGCCCTCGTAGATGG human β-actin CATGTACGTTGCTATCCAGGC CTCCTTAATGTCACGCACGAT ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 3. Primer sequences for ChIP (5’-3’) Gene promoter Forward Reverse mouse Insulin1, 2 GGAACTGTGAAACAGTCCAAGG CCCCCTGGACTTTGCTGTTTG ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 4. Primer sequences for PCR (5’-3’) Gene Forward Reverse HRD1-pDsred CCCAAGCTTATGTTCCGCACCGCAGT GGGGTACCCAGTGGGCAACAGGGG HRD1-pCMV- Flag GGGGTACCATGTTCCGCACCGCAGT CCCAAGCTTGTGGGCAACAGGGGACT C HRD1-pCMV-HA GGCCATGGGCCATATGGGATCCTTCC AGGGATGCCACCCGGGGATCCTCAGT GCACCGCAGTGATG GGGCAACAGGGGAC HRD1-N-HA GGCCATGGGCCATATGGGATCCTTCC -

Genome-Wide Analysis of Epigenetic Silencing Identifies BEX1 and BEX2 As Candidate Tumor Suppressor Genes in Malignant Glioma
Research Article Genome-Wide Analysis of Epigenetic Silencing Identifies BEX1 and BEX2 as Candidate Tumor Suppressor Genes in Malignant Glioma Greg Foltz,1,3 Gi-Yung Ryu,1 Jae-Geun Yoon,1 Timothy Nelson,1 Jessica Fahey,1 Amanda Frakes,1 Hwahyung Lee,1 Lorie Field,1 Kaitlin Zander,1 Zita Sibenaller,1 Timothy C. Ryken,1 Rajeev Vibhakar,2 Leroy Hood,3 and Anup Madan1,3 1Neurogenomic Research Laboratory, Department of Neurosurgery; 2Department of Pediatrics, University of Iowa Carver College of Medicine, Iowa City, Iowa; and 3Institute for Systems Biology, Seattle, Washington Abstract common underlying epigenetic mechanisms, such as promoter hypermethylation, histone deacetylation, histone methylation, Promoter hypermethylation and histone deacetylation are and other histone modifications, which directly or indirectly common epigenetic mechanisms implicated in the transcrip- alter chromatin structure (3–5). When combined with microarray tional silencing of tumor suppressor genes in human cancer. analysis, the pharmacologic reversal of one or more of these We treated two immortalized glioma cell lines, T98 and U87, common underlying epigenetic mechanisms provides a powerful and 10patient-derived primary glioma cell lines with screening tool for the comprehensive identification of epigenet- trichostatin A (TSA), a histone deacetylase inhibitor, or 5- ically silenced genes on a global scale. Several recent studies aza-2¶-deoxycytidine (5-AzaC), a DNA methyltransferase in- have validated this large-scale approach using DNA methyltrans- hibitor, to comprehensively identify the cohort of genes ferase and histone deacetylase (HDAC) inhibitors, either alone or reactivated through the pharmacologic reversal of these in combination, in a variety of human cancers, including distinct but related epigenetic processes. -

Proteomic Analysis of Exosome-Like Vesicles Derived from Breast Cancer Cells
ANTICANCER RESEARCH 32: 847-860 (2012) Proteomic Analysis of Exosome-like Vesicles Derived from Breast Cancer Cells GEMMA PALAZZOLO1, NADIA NINFA ALBANESE2,3, GIANLUCA DI CARA3, DANIEL GYGAX4, MARIA LETIZIA VITTORELLI3 and IDA PUCCI-MINAFRA3 1Institute for Biomedical Engineering, Laboratory of Biosensors and Bioelectronics, ETH Zurich, Switzerland; 2Department of Physics, University of Palermo, Palermo, Italy; 3Centro di Oncobiologia Sperimentale (C.OB.S.), Oncology Department La Maddalena, Palermo, Italy; 4Institute of Chemistry and Bioanalytics, University of Applied Sciences Northwestern Switzerland FHNW, Muttenz, Switzerland Abstract. Background/Aim: The phenomenon of membrane that vesicle production allows neoplastic cells to exert different vesicle-release by neoplastic cells is a growing field of interest effects, according to the possible acceptor targets. For instance, in cancer research, due to their potential role in carrying a vesicles could potentiate the malignant properties of adjacent large array of tumor antigens when secreted into the neoplastic cells or activate non-tumoral cells. Moreover, vesicles extracellular medium. In particular, experimental evidence show could convey signals to immune cells and surrounding stroma that at least some of the tumor markers detected in the blood cells. The present study may significantly contribute to the circulation of mammary carcinoma patients are carried by knowledge of the vesiculation phenomenon, which is a critical membrane-bound vesicles. Thus, biomarker research in breast device for trans cellular communication in cancer. cancer can gain great benefits from vesicle characterization. Materials and Methods: Conditioned medium was collected The phenomenon of membrane release in the extracellular from serum starved MDA-MB-231 sub-confluent cell cultures medium has long been known and was firstly described by and exosome-like vesicles (ELVs) were isolated by Paul H. -

Datasheet: VPA00601 Product Details
Datasheet: VPA00601 Description: RABBIT ANTI FKBP1A Specificity: FKBP1A Format: Purified Product Type: PrecisionAb™ Polyclonal Isotype: Polyclonal IgG Quantity: 100 µl Product Details Applications This product has been reported to work in the following applications. This information is derived from testing within our laboratories, peer-reviewed publications or personal communications from the originators. Please refer to references indicated for further information. For general protocol recommendations, please visit www.bio-rad-antibodies.com/protocols. Yes No Not Determined Suggested Dilution Western Blotting 1/1000 PrecisionAb antibodies have been extensively validated for the western blot application. The antibody has been validated at the suggested dilution. Where this product has not been tested for use in a particular technique this does not necessarily exclude its use in such procedures. Further optimization may be required dependant on sample type. Target Species Human Product Form Purified IgG - liquid Preparation Rabbit polyclonal antibody purified by affinity chromatography Buffer Solution Phosphate buffered saline Preservative 0.09% Sodium Azide (NaN3) Stabilisers 2% Sucrose Immunogen Synthetic peptide directed towards the middle region of human FKBP1A External Database Links UniProt: P62942 Related reagents Entrez Gene: 2280 FKBP1A Related reagents Synonyms FKBP1, FKBP12 Specificity Rabbit anti Human FKBP1A antibody recognizes the peptidyl-prolyl cis-trans isomerase FKBP1A, also known as 12 kDa FK506-binding protein, 12 kDa FKBP, FKBP12-Exip3, PPIase FKBP1A, calstabin 1, immunophilin FKBP12, protein kinase C inhibitor 2 or rotamase. Page 1 of 2 The protein encoded by the FKBP1A gene is a member of the immunophilin protein family, which play a role in immunoregulation and basic cellular processes involving protein folding and trafficking. -

Senescence Inhibits the Chaperone Response to Thermal Stress
SUPPLEMENTAL INFORMATION Senescence inhibits the chaperone response to thermal stress Jack Llewellyn1, 2, Venkatesh Mallikarjun1, 2, 3, Ellen Appleton1, 2, Maria Osipova1, 2, Hamish TJ Gilbert1, 2, Stephen M Richardson2, Simon J Hubbard4, 5 and Joe Swift1, 2, 5 (1) Wellcome Centre for Cell-Matrix Research, Oxford Road, Manchester, M13 9PT, UK. (2) Division of Cell Matrix Biology and Regenerative Medicine, School of Biological Sciences, Faculty of Biology, Medicine and Health, Manchester Academic Health Science Centre, University of Manchester, Manchester, M13 9PL, UK. (3) Current address: Department of Biomedical Engineering, University of Virginia, Box 800759, Health System, Charlottesville, VA, 22903, USA. (4) Division of Evolution and Genomic Sciences, School of Biological Sciences, Faculty of Biology, Medicine and Health, Manchester Academic Health Science Centre, University of Manchester, Manchester, M13 9PL, UK. (5) Correspondence to SJH ([email protected]) or JS ([email protected]). Page 1 of 11 Supplemental Information: Llewellyn et al. Chaperone stress response in senescence CONTENTS Supplemental figures S1 – S5 … … … … … … … … 3 Supplemental table S6 … … … … … … … … 10 Supplemental references … … … … … … … … 11 Page 2 of 11 Supplemental Information: Llewellyn et al. Chaperone stress response in senescence SUPPLEMENTAL FIGURES Figure S1. A EP (passage 3) LP (passage 16) 200 µm 200 µm 1.5 3 B Mass spectrometry proteomics (n = 4) C mRNA (n = 4) D 100k EP 1.0 2 p < 0.0001 p < 0.0001 LP p < 0.0001 p < 0.0001 ) 0.5 1 2 p < 0.0001 p < 0.0001 10k 0.0 0 -0.5 -1 Cell area (µm Cell area fold change vs. EP fold change vs. -

FK506-Binding Protein 12.6/1B, a Negative Regulator of [Ca<Sup>2+
University of Kentucky UKnowledge Pharmacology and Nutritional Sciences Faculty Pharmacology and Nutritional Sciences Publications Repository Citation Gant, John C.; Blalock, Eric M.; Chen, Kuey-Chu; Kadish, Inga; Thibault, Olivier; Porter, Nada M.; and Landfield, Philip W., "FK506-Binding Protein 12.6/1b, a Negative Regulator of [Ca2+], Rescues Memory and Restores Genomic Regulation in the Hippocampus of Aging Rats" (2018). Pharmacology and Nutritional Sciences Faculty Publications. 65. https://uknowledge.uky.edu/pharmacol_facpub/65 This Article is brought to you for free and open access by the Pharmacology and Nutritional Sciences at UKnowledge. It has been accepted for inclusion in Pharmacology and Nutritional Sciences Faculty Publications by an authorized administrator of UKnowledge. For more information, please contact [email protected]. 1-24-2018 FK506-Binding Protein 12.6/1b, a Negative Regulator of [Ca2+], Rescues Memory and Restores Genomic Regulation in the Hippocampus of Aging Rats John C. Gant University of Kentucky, [email protected] Eric M. Blalock University of Kentucky, [email protected] Kuey-Chu Chen University of Kentucky, [email protected] Inga Kadish University of Alabama at Birmingham Olivier Thibault University of Kentucky, [email protected] See next page for additional authors Right click to open a feedback form in a new tab to let us know how this document benefits oy u. Follow this and additional works at: https://uknowledge.uky.edu/pharmacol_facpub Part of the Cell and Developmental Biology Commons, Diseases Commons, Medical Nutrition Commons, Medical Pharmacology Commons, Neuroscience and Neurobiology Commons, and the Pharmacology, Toxicology and Environmental Health Commons Authors John C. Gant, Eric M. -

FKBP1A Sirna Set I FKBP1A Sirna Set I
Catalog # Aliquot Size F37-911-05 3 x 5 nmol F37-911-20 3 x 20 nmol F37-911-50 3 x 50 nmol FKBP1A siRNA Set I siRNA duplexes targeted against three exon regions Catalog # F37-911 Lot # Z2037-46 Specificity Formulation FKBP1A siRNAs are designed to specifically knock-down The siRNAs are supplied as a lyophilized powder and human FKBP1A expression. shipped at room temperature. Product Description Reconstitution Protocol FKBP1A siRNA is a pool of three individual synthetic siRNA Briefly centrifuge the tubes (maximum RCF 4,000g) to duplexes designed to knock-down human FKBP1A mRNA collect lyophilized siRNA at the bottom of the tube. expression. Each siRNA is 19-25 bases in length. The gene Resuspend the siRNA in 50 µl of DEPC-treated water accession number is NM_000801. (supplied by researcher), which results in a 1x stock solution (10 µM). Gently pipet the solution 3-5 times to mix Gene Aliases and avoid the introduction of bubbles. Optional: aliquot FKBP12, FKBP1, PKC12, PKCI2, PPIASE, FKBP-12, FKBP12C 1x stock solutions for storage. Storage and Stability Related Products The lyophilized powder is stable for at least 4 weeks at room temperature. It is recommended that the Product Name Catalog Number lyophilized and resuspended siRNAs are stored at or FKBP1A Protein F37-30G below -20oC. After resuspension, siRNA stock solutions ≥2 µM can undergo up to 50 freeze-thaw cycles without significant degradation. For long-term storage, it is recommended that the siRNA is stored at -70oC. For most favorable performance, avoid repeated handling and multiple freeze/thaw cycles. -

FKBP1A Protein FKBP1A Protein
Catalogue # Aliquot Size F37-30G-50 50 µg F37-30G-200 200 µg FKBP1A Protein Full length recombinant protein expressed in E.coli cells Catalog # F37-30G Lot # W140 -5 Product Description Purity Recombinant full length human FKBP1A was expressed in E. coli cells using an N-terminal GST tag. The gene accession number is NM_000801 . The purity was determined to be Gene Aliases >95% by densitometry. Approx. MW 38.3kDa . FKBP12, FKBP1, PKC12, PKCI2, PPIASE, FKBP-12, FKBP12C Formulation Recombinant protein stored in 50mM Tris-HCl, pH 7.5, 150mM NaCl, 10mM glutathione, 0.1mM EDTA, 0.25mM DTT, 0.1mM PMSF, 25% glycerol. Storage and Stability o Store product at –70 C. For optimal storage, aliquot target into smaller quantities after centrifugation and store at recommended temperature. For most favorable performance, avoid repeated handling and multiple freeze/thaw cycles. Scientific Background FKBP1A is a member of the immunophilin protein family which plays a role in immunoregulation and basic cellular processes involving protein folding and trafficking. FKBP1A is a cis-trans prolyl isomerase that binds the FKBP1A Protein immunosuppressants FK506 and Rapamycin. FKBP1A Full length recombinant protein expressed in E. coli cells interacts with various type I receptors, including the TGF- beta type I receptor and competitive binding assays Catalog Number F37-30G show that the type I receptor may be a natural ligand for Specific Lot Number W140-5 FKBP1A (1). FKBP1A-deficient mice have normal skeletal Purity >95% muscle but show severe dilated cardiomyopathy and Concentration 0.2µg/ µl ventricular septal defects that mimick a human Stability 1yr at –70 oC from date of shipment Storage & Shipping Store product at –70 oC.