Supplementary Table 1. Unique Proteins with Expression Significantly Altered in the Hippocampus of Rats of Group of Exposed to Mehg Group (EG) Vs
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ATP-Induced Focal Adhesion Kinase Activity Is Negatively Modulated by Phospholipase D2 in PC12 Cells
EXPERIMENTAL and MOLECULAR MEDICINE, Vol. 33, No. 3, 150-155, September 2001 ATP-induced focal adhesion kinase activity is negatively modulated by phospholipase D2 in PC12 cells Yoe-Sik Bae1 and Sung Ho Ryu1,2 Introduction 1 Division of Molecular and Life Sciences, Pohang University of Purinergic receptors have been reported to play impor- Science and Technology, Pohang 790-784, Korea tant roles on the regulation of neuronal cell functions 2 Corresponding author: Tel, +82-54-279-2292; (Communi et al., 2000; Di Iorio et al., 1998). ATP, a Fax, +82-54-279-2199; E-mail, [email protected] ligand for the receptors modulate various cellular re- sponses such as mitogenic and morphogenic activity in Accepted 18 September 2001 PC12 rat pheochromocytoma cells (Neary et al., 1996; Soltoff et al., 1998; Schindelholz et al., 2000). Stimu- Abbreviations: Fak, focal adhesion kinase; PLD, phospholipase D; lation of cells with ATP induces tyrosine phosphorylation PA, phosphatidic acid; PC, phosphatidylcholine; DAG, diacylglyc- of several cytoskeletal proteins and focal adhesion erol; PBt, phosphatidylbutanol; PKC, protein kinase C; PAP, phos- molecules such as focal adhesion kinase (Fak), proline- phatidic acid phosphohydrolase rich tyrosine kinase (Pyk2), and paxillin (Soltoff et al., 1998; Schindelholz et al., 2000). Since these cytosk- eleton-associated proteins have been regarded as important factors for the regulation of neuronal cell Abstract functions, the study on the regulatory mechanism for the proteins remains an important issue. Extracellular ATP has been known to modulate vari- Phospholipase D (PLD) catalyzes the hydrolysis of ous cellular responses including mitogenesis, secre- phosphatidylcholine (PC) into phosphatidic acid (PA) tion and morphogenic activity in neuronal cells. -

Proteomic Analyses Reveal a Role of Cytoplasmic Droplets As an Energy Source During Sperm Epididymal Maturation
Proteomic analyses reveal a role of cytoplasmic droplets as an energy source during sperm epididymal maturation Shuiqiao Yuana,b, Huili Zhenga, Zhihong Zhengb, Wei Yana,1 aDepartment of Physiology and Cell Biology, University of Nevada School of Medicine, Reno, NV, 89557; and bDepartment of Laboratory Animal Medicine, China Medical University, Shenyang, 110001, China Corresponding author. Email: [email protected] Supplemental Information contains one Figure (Figure S1), three Tables (Tables S1-S3) and two Videos (Videos S1 and S2) files. Figure S1. Scanning electron microscopic images of purified murine cytoplasmic droplets. Arrows point to indentations resembling the resealed defects at the detaching points when CDs come off the sperm flagella. Scale bar = 1µm Table S1 Mass spectrometry-based identifiaction of proteins highly enriched in murine cytoplasmic droplets. # MS/MS View:Identified Proteins (105) Accession Number Molecular Weight Protein Grouping Ambiguity Dot_1_1 Dot_2_1 Dot_3_1 Dot_4_1Dot_5_1 Dot_1_2 Dot_2_2 Dot_3_2 Dot_4_2 Dot_5_2 1 IPI:IPI00467457.3 Tax_Id=10090 Gene_Symbol=Ldhc L-lactate dehydrogenase C chain IPI00467457 36 kDa TRUE 91% 100% 100% 100% 100% 100% 100% 100% 100% 2 IPI:IPI00473320.2 Tax_Id=10090 Gene_Symbol=Actb Putative uncharacterized protein IPI00473320 42 kDa TRUE 75% 100% 100% 100% 100% 89% 76% 100% 100% 100% 3 IPI:IPI00224181.7 Tax_Id=10090 Gene_Symbol=Akr1b7 Aldose reductase-related protein 1 IPI00224181 36 kDa TRUE 100% 100% 76% 100% 100% 4 IPI:IPI00228633.7 Tax_Id=10090 Gene_Symbol=Gpi1 Glucose-6-phosphate -

Datasheet for Protein Phosphatase 1 (PP1) (P0754; Lot 0121306)
Supplied in: 200 mM NaCl, 50 mM HEPES Unit Definition: One unit is defined as Notes On Use: Avoid freeze/thaw cycles. Can be Protein the amount of enzyme that hydrolyzes 1 nmol of stored for 1 week or less at –20°C. (pH 7.0 @ 25°C), 1 mM MnCl2, 0.1 mM EGTA, Phosphatase 1 2.5 mM dithiothreitol, 0.025% Tween-20 and p-Nitrophenyl Phosphate (50 mM) (NEB #P0757) 50% glycerol. Store at –70°C in 1 minute at 30°C in a total reaction volume of The following information can be used as (PP1) 50 µl. suggested initial conditions for dephosphorylation 1-800-632-7799 Applications: PP1 can be used to release of proteins with PP1. [email protected] phosphate groups from phosphorylated serine, Specific Activity: ~ 80,000 units/mg. www.neb.com 0.1 unit of PP1 removes ~100% of phosphates P0754S 012130614061 threonine and tyrosine residues in proteins. Note that different proteins are dephosphorylated at Molecular Weight: 37.5 kDa. (0.5 nmol) from phosphoserine/threonine different rates. residues in phosphorylase a as well as in P0754S r y Purity: PP1 has been purified to > 90% phosphorylated myelin basic protein (phospho- 100 units 2,500 U/ml Lot: 0121306 Reagents Supplied with Enzyme: homogeneity as determined by SDS-PAGE and MyBP, 18.5 kDa) in 30 minutes in a 50 µl reaction. RECOMBINANT Store at –70°C Exp: 6/14 10X NEBuffer for Protein MetalloPhosphatases Coomassie Blue staining. The concentration of phospho-MyBP is 10 µM (PMP) with respect to phosphate. Description: Protein Phosphatase 1 (PP1) is a 10X MnCl2 (10 mM) Quality Assurance: PP1 contains no detectable Mn2+-dependent protein phosphatase with activity protease activity. -

Role of Phospholipases in Adrenal Steroidogenesis
229 1 W B BOLLAG Phospholipases in adrenal 229:1 R29–R41 Review steroidogenesis Role of phospholipases in adrenal steroidogenesis Wendy B Bollag Correspondence should be addressed Charlie Norwood VA Medical Center, One Freedom Way, Augusta, GA, USA to W B Bollag Department of Physiology, Medical College of Georgia, Augusta University (formerly Georgia Regents Email University), Augusta, GA, USA [email protected] Abstract Phospholipases are lipid-metabolizing enzymes that hydrolyze phospholipids. In some Key Words cases, their activity results in remodeling of lipids and/or allows the synthesis of other f adrenal cortex lipids. In other cases, however, and of interest to the topic of adrenal steroidogenesis, f angiotensin phospholipases produce second messengers that modify the function of a cell. In this f intracellular signaling review, the enzymatic reactions, products, and effectors of three phospholipases, f phospholipids phospholipase C, phospholipase D, and phospholipase A2, are discussed. Although f signal transduction much data have been obtained concerning the role of phospholipases C and D in regulating adrenal steroid hormone production, there are still many gaps in our knowledge. Furthermore, little is known about the involvement of phospholipase A2, Endocrinology perhaps, in part, because this enzyme comprises a large family of related enzymes of that are differentially regulated and with different functions. This review presents the evidence supporting the role of each of these phospholipases in steroidogenesis in the Journal Journal of Endocrinology adrenal cortex. (2016) 229, R1–R13 Introduction associated GTP-binding protein exchanges a bound GDP for a GTP. The G protein with GTP bound can then Phospholipids serve a structural function in the cell in that activate the enzyme, phospholipase C (PLC), that cleaves they form the lipid bilayer that maintains cell integrity. -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of -
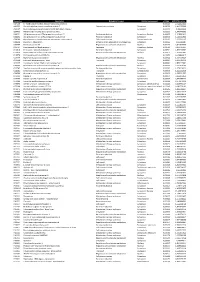
Accession Description Biological Process Cellular Component P-Value SCZ/CTRL Ration A4FU69 EF-Hand Calcium-Binding Domain-Contai
Accession Description Biological Process Cellular component p-Value SCZ/CTRL ration A4FU69 EF-hand calcium-binding domain-containing protein 5 0,001101 2,724427411 A4UGR9 Xin actin-binding repeat-containing protein 2 Cytoskeletal anchoring Cytoplasm 0,006756 1,413953388 A6NCE7 Microtubule-associated proteins 1A/1B light chain 3 beta 2 Cytoplasm 0,001417 1,99612969 B2RPK0 Putative high mobility group protein B1-like 1 0,032314 1,590743352 O00231 26S proteasome non-ATPase regulatory subunit 11 Protein metabolism Cytoplasm; Nucleus 0,000029 1,428664042 O00232 26S proteasome non-ATPase regulatory subunit 12 Protein metabolism Cytoplasm 0,008566 1,544545922 O00264 Membrane-associated progesterone receptor component 1 Cell communication Plasma membrane 0,001459 2,322924147 O00429 Dynamin-1-like protein Mitochondrion organization and biogenesis Cytoplasm 0,006560 0,06391487 O00567 Nucleolar protein 56 Regulation of nucleotide metabolism Nucleus 0,007330 3,842896338 O14737 Programmed cell death protein 5 Apoptosis Cytoplasm; Nucleus 0,006358 3,836727237 O14818 Proteasome subunit alpha type-7 Protein metabolism Cytoplasm 0,030521 1,893928387 O14979 Heterogeneous nuclear ribonucleoprotein D-like Regulation of nucleotide metabolism Nucleus 0,000637 2,150005885 O15078 Centrosomal protein of 290 kDa 0,015359 14,26648619 O15347 High mobility group protein B3 Regulation of nucleotide metabolism Nucleus 0,005500 1,364309014 O15540 Fatty acid-binding protein_ brain Transport Cytoplasm 0,000087 3,125786118 O43237 Cytoplasmic dynein 1 light intermediate -

The Regulatory Roles of Phosphatases in Cancer
Oncogene (2014) 33, 939–953 & 2014 Macmillan Publishers Limited All rights reserved 0950-9232/14 www.nature.com/onc REVIEW The regulatory roles of phosphatases in cancer J Stebbing1, LC Lit1, H Zhang, RS Darrington, O Melaiu, B Rudraraju and G Giamas The relevance of potentially reversible post-translational modifications required for controlling cellular processes in cancer is one of the most thriving arenas of cellular and molecular biology. Any alteration in the balanced equilibrium between kinases and phosphatases may result in development and progression of various diseases, including different types of cancer, though phosphatases are relatively under-studied. Loss of phosphatases such as PTEN (phosphatase and tensin homologue deleted on chromosome 10), a known tumour suppressor, across tumour types lends credence to the development of phosphatidylinositol 3--kinase inhibitors alongside the use of phosphatase expression as a biomarker, though phase 3 trial data are lacking. In this review, we give an updated report on phosphatase dysregulation linked to organ-specific malignancies. Oncogene (2014) 33, 939–953; doi:10.1038/onc.2013.80; published online 18 March 2013 Keywords: cancer; phosphatases; solid tumours GASTROINTESTINAL MALIGNANCIES abs in sera were significantly associated with poor survival in Oesophageal cancer advanced ESCC, suggesting that they may have a clinical utility in Loss of PTEN (phosphatase and tensin homologue deleted on ESCC screening and diagnosis.5 chromosome 10) expression in oesophageal cancer is frequent, Cao et al.6 investigated the role of protein tyrosine phosphatase, among other gene alterations characterizing this disease. Zhou non-receptor type 12 (PTPN12) in ESCC and showed that PTPN12 et al.1 found that overexpression of PTEN suppresses growth and protein expression is higher in normal para-cancerous tissues than induces apoptosis in oesophageal cancer cell lines, through in 20 ESCC tissues. -

Inhibition of Protein Phosphatase 1 Translation by Preventing JNK
Active ERK Contributes to Protein Translation by Preventing JNK-Dependent Inhibition of Protein Phosphatase 1 This information is current as Martha M. Monick, Linda S. Powers, Thomas J. Gross, of September 28, 2021. Dawn M. Flaherty, Christopher W. Barrett and Gary W. Hunninghake J Immunol 2006; 177:1636-1645; ; doi: 10.4049/jimmunol.177.3.1636 http://www.jimmunol.org/content/177/3/1636 Downloaded from References This article cites 59 articles, 43 of which you can access for free at: http://www.jimmunol.org/content/177/3/1636.full#ref-list-1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication by guest on September 28, 2021 *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2006 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Active ERK Contributes to Protein Translation by Preventing JNK-Dependent Inhibition of Protein Phosphatase 11 Martha M. Monick,2 Linda S. Powers, Thomas J. Gross, Dawn M. Flaherty, Christopher W. -

Looking for Missing Proteins in the Proteome Of
Looking for Missing Proteins in the Proteome of Human Spermatozoa: An Update Yves Vandenbrouck, Lydie Lane, Christine Carapito, Paula Duek, Karine Rondel, Christophe Bruley, Charlotte Macron, Anne Gonzalez de Peredo, Yohann Coute, Karima Chaoui, et al. To cite this version: Yves Vandenbrouck, Lydie Lane, Christine Carapito, Paula Duek, Karine Rondel, et al.. Looking for Missing Proteins in the Proteome of Human Spermatozoa: An Update. Journal of Proteome Research, American Chemical Society, 2016, 15 (11), pp.3998-4019. 10.1021/acs.jproteome.6b00400. hal-02191502 HAL Id: hal-02191502 https://hal.archives-ouvertes.fr/hal-02191502 Submitted on 19 Mar 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Journal of Proteome Research 1 2 3 Looking for missing proteins in the proteome of human spermatozoa: an 4 update 5 6 Yves Vandenbrouck1,2,3,#,§, Lydie Lane4,5,#, Christine Carapito6, Paula Duek5, Karine Rondel7, 7 Christophe Bruley1,2,3, Charlotte Macron6, Anne Gonzalez de Peredo8, Yohann Couté1,2,3, 8 Karima Chaoui8, Emmanuelle Com7, Alain Gateau5, AnneMarie Hesse1,2,3, Marlene 9 Marcellin8, Loren Méar7, Emmanuelle MoutonBarbosa8, Thibault Robin9, Odile Burlet- 10 Schiltz8, Sarah Cianferani6, Myriam Ferro1,2,3, Thomas Fréour10,11, Cecilia Lindskog12,Jérôme 11 1,2,3 7,§ 12 Garin , Charles Pineau . -

Supplementary Data Genbank Or OSE Vs RO NIA Accession Gene Name Symbol FC B-Value H3073C09 11.38 5.62 H3126B09 9.64 6.44 H3073B0
Supplementary Data GenBank or OSE vs RO NIA accession Gene name Symbol FC B-value H3073C09 11.38 5.62 H3126B09 9.64 6.44 H3073B08 9.62 5.59 AU022767 Exportin 4 Xpo4 9.62 6.64 H3073B09 9.59 6.48 BG063925 Metallothionein 2 Mt2 9.23 18.89 H3064B07 9.21 6.10 H3073D08 8.28 6.10 AU021923 Jagged 1 Jag1 7.89 5.93 H3070D08 7.54 4.58 BG085110 Cysteine-rich protein 1 (intestinal) Crip1 6.23 16.40 BG063004 Lectin, galactose binding, soluble 1 Lgals1 5.95 10.36 BG069712 5.92 2.34 BG076976 Transcribed locus, strongly similar to NP_032521.1 lectin, galactose binding, soluble 1 5.64 8.36 BG062930 DNA segment, Chr 11, Wayne State University 99, expressed D11Wsu99e 5.63 8.76 BG086474 Insulin-like growth factor binding protein 5 Igfbp5 5.50 15.95 H3002d11 5.13 20.77 BG064706 Keratin complex 1, acidic, gene 19 Krt1-19 5.06 9.07 H3007A09 5.05 2.46 H3065F02 4.84 5.43 BG081752 4.81 1.25 H3010E09 4.71 11.90 H3064c11 4.43 1.00 BG069711 Transmembrane 4 superfamily member 9 Tm4sf9 4.29 1.23 BG077072 Actin, beta, cytoplasmic Actb 4.29 3.01 BG079788 Hemoglobin alpha, adult chain 1 Hba-a1 4.26 6.63 BG076798 4.23 0.80 BG074344 Mesothelin Msln 4.22 6.97 C78835 Actin, beta, cytoplasmic Actb 4.16 3.02 BG067531 4.15 1.61 BG073468 Hemoglobin alpha, adult chain 1 Hba-a1 4.10 6.23 H3154H07 4.08 5.38 AW550167 3.95 5.66 H3121B01 3.94 5.94 H3124f12 3.94 5.64 BG073608 Hemoglobin alpha, adult chain 1 Hba-a1 3.84 5.32 BG073617 Hemoglobin alpha, adult chain 1 Hba-a1 3.84 5.75 BG072574 Hemoglobin alpha, adult chain 1 Hba-a1 3.82 5.93 BG072211 Tumor necrosis factor receptor superfamily, -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -

CX3CR1 Engagement by Respiratory Syncytial Virus Leads to Induction of Nucleolin And
bioRxiv preprint doi: https://doi.org/10.1101/2020.07.29.227967; this version posted July 30, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 RESEARCH ARTICLE 2 3 CX3CR1 Engagement by Respiratory Syncytial Virus Leads to Induction of Nucleolin and 4 Dysregulation of Cilia-related Genes 5 6 Christopher S. Andersona*, Tatiana Chirkovab*, Christopher G. Slaunwhite, Xing Qiuc, Edward E. Walshd, 7 Larry J. Andersonb# and Thomas J. Mariania# 8 9 aDepartments of Pediatrics, University of Rochester Medical Center, Rochester, NY. 10 bEmory University Department of Pediatrics and Children’s Healthcare of Atlanta, Atlanta, GA. 11 cDepartment of Biostatistics and Computational Biology 12 dDepartment of Medicine, University of Rochester Medical Center, Rochester, NY. 13 *contributed equally to this work 14 #corresponding authors 15 16 Address for Correspondence: 17 Thomas J. Mariani, PhD 18 Professor of Pediatrics 19 Division of Neonatology and 20 Pediatric Molecular and Personalized Medicine Program 21 University of Rochester Medical Center 22 601 Elmwood Ave, Box 850 23 Rochester, NY 14642, USA. 24 Phone: 585-276-4616; Fax: 585-276-2643; 25 E-mail: [email protected] 26 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.07.29.227967; this version posted July 30, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity.