Cell-Specific Bioorthogonal Tagging of Glycoproteins in Co- Culture
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ATP-Induced Focal Adhesion Kinase Activity Is Negatively Modulated by Phospholipase D2 in PC12 Cells
EXPERIMENTAL and MOLECULAR MEDICINE, Vol. 33, No. 3, 150-155, September 2001 ATP-induced focal adhesion kinase activity is negatively modulated by phospholipase D2 in PC12 cells Yoe-Sik Bae1 and Sung Ho Ryu1,2 Introduction 1 Division of Molecular and Life Sciences, Pohang University of Purinergic receptors have been reported to play impor- Science and Technology, Pohang 790-784, Korea tant roles on the regulation of neuronal cell functions 2 Corresponding author: Tel, +82-54-279-2292; (Communi et al., 2000; Di Iorio et al., 1998). ATP, a Fax, +82-54-279-2199; E-mail, [email protected] ligand for the receptors modulate various cellular re- sponses such as mitogenic and morphogenic activity in Accepted 18 September 2001 PC12 rat pheochromocytoma cells (Neary et al., 1996; Soltoff et al., 1998; Schindelholz et al., 2000). Stimu- Abbreviations: Fak, focal adhesion kinase; PLD, phospholipase D; lation of cells with ATP induces tyrosine phosphorylation PA, phosphatidic acid; PC, phosphatidylcholine; DAG, diacylglyc- of several cytoskeletal proteins and focal adhesion erol; PBt, phosphatidylbutanol; PKC, protein kinase C; PAP, phos- molecules such as focal adhesion kinase (Fak), proline- phatidic acid phosphohydrolase rich tyrosine kinase (Pyk2), and paxillin (Soltoff et al., 1998; Schindelholz et al., 2000). Since these cytosk- eleton-associated proteins have been regarded as important factors for the regulation of neuronal cell Abstract functions, the study on the regulatory mechanism for the proteins remains an important issue. Extracellular ATP has been known to modulate vari- Phospholipase D (PLD) catalyzes the hydrolysis of ous cellular responses including mitogenesis, secre- phosphatidylcholine (PC) into phosphatidic acid (PA) tion and morphogenic activity in neuronal cells. -

Datasheet for Protein Phosphatase 1 (PP1) (P0754; Lot 0121306)
Supplied in: 200 mM NaCl, 50 mM HEPES Unit Definition: One unit is defined as Notes On Use: Avoid freeze/thaw cycles. Can be Protein the amount of enzyme that hydrolyzes 1 nmol of stored for 1 week or less at –20°C. (pH 7.0 @ 25°C), 1 mM MnCl2, 0.1 mM EGTA, Phosphatase 1 2.5 mM dithiothreitol, 0.025% Tween-20 and p-Nitrophenyl Phosphate (50 mM) (NEB #P0757) 50% glycerol. Store at –70°C in 1 minute at 30°C in a total reaction volume of The following information can be used as (PP1) 50 µl. suggested initial conditions for dephosphorylation 1-800-632-7799 Applications: PP1 can be used to release of proteins with PP1. [email protected] phosphate groups from phosphorylated serine, Specific Activity: ~ 80,000 units/mg. www.neb.com 0.1 unit of PP1 removes ~100% of phosphates P0754S 012130614061 threonine and tyrosine residues in proteins. Note that different proteins are dephosphorylated at Molecular Weight: 37.5 kDa. (0.5 nmol) from phosphoserine/threonine different rates. residues in phosphorylase a as well as in P0754S r y Purity: PP1 has been purified to > 90% phosphorylated myelin basic protein (phospho- 100 units 2,500 U/ml Lot: 0121306 Reagents Supplied with Enzyme: homogeneity as determined by SDS-PAGE and MyBP, 18.5 kDa) in 30 minutes in a 50 µl reaction. RECOMBINANT Store at –70°C Exp: 6/14 10X NEBuffer for Protein MetalloPhosphatases Coomassie Blue staining. The concentration of phospho-MyBP is 10 µM (PMP) with respect to phosphate. Description: Protein Phosphatase 1 (PP1) is a 10X MnCl2 (10 mM) Quality Assurance: PP1 contains no detectable Mn2+-dependent protein phosphatase with activity protease activity. -

Propranolol-Mediated Attenuation of MMP-9 Excretion in Infants with Hemangiomas
Supplementary Online Content Thaivalappil S, Bauman N, Saieg A, Movius E, Brown KJ, Preciado D. Propranolol-mediated attenuation of MMP-9 excretion in infants with hemangiomas. JAMA Otolaryngol Head Neck Surg. doi:10.1001/jamaoto.2013.4773 eTable. List of All of the Proteins Identified by Proteomics This supplementary material has been provided by the authors to give readers additional information about their work. © 2013 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 10/01/2021 eTable. List of All of the Proteins Identified by Proteomics Protein Name Prop 12 mo/4 Pred 12 mo/4 Δ Prop to Pred mo mo Myeloperoxidase OS=Homo sapiens GN=MPO 26.00 143.00 ‐117.00 Lactotransferrin OS=Homo sapiens GN=LTF 114.00 205.50 ‐91.50 Matrix metalloproteinase‐9 OS=Homo sapiens GN=MMP9 5.00 36.00 ‐31.00 Neutrophil elastase OS=Homo sapiens GN=ELANE 24.00 48.00 ‐24.00 Bleomycin hydrolase OS=Homo sapiens GN=BLMH 3.00 25.00 ‐22.00 CAP7_HUMAN Azurocidin OS=Homo sapiens GN=AZU1 PE=1 SV=3 4.00 26.00 ‐22.00 S10A8_HUMAN Protein S100‐A8 OS=Homo sapiens GN=S100A8 PE=1 14.67 30.50 ‐15.83 SV=1 IL1F9_HUMAN Interleukin‐1 family member 9 OS=Homo sapiens 1.00 15.00 ‐14.00 GN=IL1F9 PE=1 SV=1 MUC5B_HUMAN Mucin‐5B OS=Homo sapiens GN=MUC5B PE=1 SV=3 2.00 14.00 ‐12.00 MUC4_HUMAN Mucin‐4 OS=Homo sapiens GN=MUC4 PE=1 SV=3 1.00 12.00 ‐11.00 HRG_HUMAN Histidine‐rich glycoprotein OS=Homo sapiens GN=HRG 1.00 12.00 ‐11.00 PE=1 SV=1 TKT_HUMAN Transketolase OS=Homo sapiens GN=TKT PE=1 SV=3 17.00 28.00 ‐11.00 CATG_HUMAN Cathepsin G OS=Homo -

Novel Missense Mutation in PTPN22 in a Chinese Pedigree With
Gong et al. BMC Endocrine Disorders (2018) 18:76 https://doi.org/10.1186/s12902-018-0305-8 RESEARCHARTICLE Open Access Novel missense mutation in PTPN22 in a Chinese pedigree with Hashimoto’s thyroiditis Licheng Gong1†, Beihong Liu2,3†, Jing Wang4, Hong Pan3, Anhui Qi2,3, Siyang Zhang2,3, Jinyi Wu1, Ping Yang1* and Binbin Wang3,4,5* Abstract Background: Hashimoto’s thyroiditis is a complex autoimmune thyroid disease, the onset of which is associated with environmental exposures and specific susceptibility genes. Its incidence in females is higher than its incidence in males. Thus far, although some susceptibility loci have been elaborated, including PTPN22, FOXP3, and CD25, the aetiology and pathogenesis of Hashimoto’s thyroiditis remains unclear. Methods: Four affected members from a Chinese family with Hashimoto’s thyroiditis were selected for whole-exome sequencing. Missense, nonsense, frameshift, or splicing-site variants shared by all affected members were identified after frequency filtering against public and internal exome databases. Segregation analysis was performed by Sanger sequencing among all members with available DNA. Results: We identified a missense mutation in PTPN22 (NM_015967.5; c. 77A > G; p.Asn26Ser) using whole-exome sequencing. PTPN22 is a known susceptibility gene associated with increased risks of multiple autoimmune diseases. Cosegregation analysis confirmed that all patients in this family, all of whom were female, carried the mutation. All public and private databases showed that the missense mutation was extremely rare. Conclusions: We found a missense mutation in PTPN22 in a Chinese HT pedigree using whole-exome sequencing. Our study, for the first time, linked a rare variant of PTPN22 to Hashimoto’s thyroiditis, providing further evidence of the disease-causing or susceptibility role of PTPN22 in autoimmune thyroid disease. -

Role of Phospholipases in Adrenal Steroidogenesis
229 1 W B BOLLAG Phospholipases in adrenal 229:1 R29–R41 Review steroidogenesis Role of phospholipases in adrenal steroidogenesis Wendy B Bollag Correspondence should be addressed Charlie Norwood VA Medical Center, One Freedom Way, Augusta, GA, USA to W B Bollag Department of Physiology, Medical College of Georgia, Augusta University (formerly Georgia Regents Email University), Augusta, GA, USA [email protected] Abstract Phospholipases are lipid-metabolizing enzymes that hydrolyze phospholipids. In some Key Words cases, their activity results in remodeling of lipids and/or allows the synthesis of other f adrenal cortex lipids. In other cases, however, and of interest to the topic of adrenal steroidogenesis, f angiotensin phospholipases produce second messengers that modify the function of a cell. In this f intracellular signaling review, the enzymatic reactions, products, and effectors of three phospholipases, f phospholipids phospholipase C, phospholipase D, and phospholipase A2, are discussed. Although f signal transduction much data have been obtained concerning the role of phospholipases C and D in regulating adrenal steroid hormone production, there are still many gaps in our knowledge. Furthermore, little is known about the involvement of phospholipase A2, Endocrinology perhaps, in part, because this enzyme comprises a large family of related enzymes of that are differentially regulated and with different functions. This review presents the evidence supporting the role of each of these phospholipases in steroidogenesis in the Journal Journal of Endocrinology adrenal cortex. (2016) 229, R1–R13 Introduction associated GTP-binding protein exchanges a bound GDP for a GTP. The G protein with GTP bound can then Phospholipids serve a structural function in the cell in that activate the enzyme, phospholipase C (PLC), that cleaves they form the lipid bilayer that maintains cell integrity. -

The Intrinsically Disordered Proteins of Myelin in Health and Disease
cells Review Flexible Players within the Sheaths: The Intrinsically Disordered Proteins of Myelin in Health and Disease Arne Raasakka 1 and Petri Kursula 1,2,* 1 Department of Biomedicine, University of Bergen, Jonas Lies vei 91, NO-5009 Bergen, Norway; [email protected] 2 Faculty of Biochemistry and Molecular Medicine & Biocenter Oulu, University of Oulu, Aapistie 7A, FI-90220 Oulu, Finland * Correspondence: [email protected] Received: 30 January 2020; Accepted: 16 February 2020; Published: 18 February 2020 Abstract: Myelin ensheathes selected axonal segments within the nervous system, resulting primarily in nerve impulse acceleration, as well as mechanical and trophic support for neurons. In the central and peripheral nervous systems, various proteins that contribute to the formation and stability of myelin are present, which also harbor pathophysiological roles in myelin disease. Many myelin proteins have common attributes, including small size, hydrophobic segments, multifunctionality, longevity, and regions of intrinsic disorder. With recent advances in protein biophysical characterization and bioinformatics, it has become evident that intrinsically disordered proteins (IDPs) are abundant in myelin, and their flexible nature enables multifunctionality. Here, we review known myelin IDPs, their conservation, molecular characteristics and functions, and their disease relevance, along with open questions and speculations. We place emphasis on classifying the molecular details of IDPs in myelin, and we correlate these with their various functions, including susceptibility to post-translational modifications, function in protein–protein and protein–membrane interactions, as well as their role as extended entropic chains. We discuss how myelin pathology can relate to IDPs and which molecular factors are potentially involved. Keywords: myelin; intrinsically disordered protein; multiple sclerosis; peripheral neuropathies; myelination; protein folding; protein–membrane interaction; protein–protein interaction 1. -

Ɑ6ß1 Andɑ7ß1 Integrins Are Required in Schwann Cells to Sort
The Journal of Neuroscience, November 13, 2013 • 33(46):17995–18007 • 17995 Cellular/Molecular ␣61 and ␣71 Integrins Are Required in Schwann Cells to Sort Axons Marta Pellegatta,1,2 Ade`le De Arcangelis,3 Alessandra D’Urso,1 Alessandro Nodari,1 Desire´e Zambroni,1 Monica Ghidinelli,1,2 Vittoria Matafora,1 Courtney Williamson,2 Elisabeth Georges-Labouesse,3† Jordan Kreidberg,4 Ulrike Mayer,5 Karen K. McKee,6 Peter D. Yurchenco,6 Angelo Quattrini,1 Lawrence Wrabetz,1,2 and Maria Laura Feltri1,2 1San Raffaele Scientific Institute, Milano 20132, Italy, 2Hunter James Kelly Research Institute, University at Buffalo, State University of New York, New York 14203, 3Development and Stem Cells Program, Institut de Ge´ne´tique et de Biologie Mole´culaire et Cellulaire, Centre National de la Recherche Scientifique, Unite´ Mixte de Recherche 7104, Institut National de la Sante´ et de la Recherche Me´dicale U964, Universite´ de Strasbourg, Illkirch 67404, France, 4Department of Medicine, Children’s Hospital Boston and Department of Pediatrics, Harvard Medical School, Boston, Massachusetts 02115, 5Biomedical Research Centre, School of Biological Sciences, University of East Anglia, Norwich NR4 7TJ, United Kingdom, and 6Robert Wood Johnson Medical School, Piscataway, New Jersey, New Jersey 08854 During development, Schwann cells extend lamellipodia-like processes to segregate large- and small-caliber axons during the process of radial sorting. Radial sorting is a prerequisite for myelination and is arrested in human neuropathies because of laminin deficiency. Experiments in mice using targeted mutagenesis have confirmed that laminins 211, 411, and receptors containing the 1 integrin subunit are required for radial sorting; however, which of the 11 ␣ integrins that can pair with 1 forms the functional receptor is unknown. -

Open Full Page
Published OnlineFirst December 17, 2015; DOI: 10.1158/0008-5472.CAN-15-0884 Cancer Tumor and Stem Cell Biology Research Eva1 Maintains the Stem-like Character of Glioblastoma-Initiating Cells by Activating the Noncanonical NF-kB Signaling Pathway Naoki Ohtsu1, Yuka Nakatani2, Daisuke Yamashita3, Shiro Ohue3, Takanori Ohnishi3,and Toru Kondo1,2 Abstract Glioblastoma (GBM)–initiating cells (GIC) are a tumorigenic as Eva1 overexpression enhanced these properties. Eva1 deficien- subpopulation that are resistant to radio- and chemotherapies cy was also associated with decreased expression of stemness- and are the source of disease recurrence. Therefore, the identifi- related genes, indicating a requirement for Eva1 in maintaining cation and characterization of GIC-specific factors is critical GIC pluripotency. We further demonstrate that Eva1 induced GIC toward the generation of effective GBM therapeutics. In this study, proliferation through the activation of the RelB-dependent non- we investigated the role of epithelial V-like antigen 1 (Eva1, also canonical NF-kB pathway by recruiting TRAF2 to the cytoplasmic known as myelin protein zero-like 2) in stemness and GBM tail. Taken together, our findings highlight Eva1 as a novel tumorigenesis. Eva1 was prominently expressed in GICs in vitro regulator of GIC function and also provide new mechanistic and in stem cell marker (Sox2, CD15, CD49f)-expressing cells insight into the role of noncanonical NF-kB activation in GIC, derived from human GBM tissues. Eva1 knockdown in GICs thus offering multiple potential therapeutic targets for preclinical reduced their self-renewal and tumor-forming capabilities, where- investigation in GBM. Cancer Res; 76(1); 171–81. Ó2015 AACR. -

How Does Protein Zero Assemble Compact Myelin?
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 13 May 2020 doi:10.20944/preprints202005.0222.v1 Peer-reviewed version available at Cells 2020, 9, 1832; doi:10.3390/cells9081832 Perspective How Does Protein Zero Assemble Compact Myelin? Arne Raasakka 1,* and Petri Kursula 1,2 1 Department of Biomedicine, University of Bergen, Jonas Lies vei 91, NO-5009 Bergen, Norway 2 Faculty of Biochemistry and Molecular Medicine & Biocenter Oulu, University of Oulu, Aapistie 7A, FI-90220 Oulu, Finland; [email protected] * Correspondence: [email protected] Abstract: Myelin protein zero (P0), a type I transmembrane protein, is the most abundant protein in peripheral nervous system (PNS) myelin – the lipid-rich, periodic structure that concentrically encloses long axonal segments. Schwann cells, the myelinating glia of the PNS, express P0 throughout their development until the formation of mature myelin. In the intramyelinic compartment, the immunoglobulin-like domain of P0 bridges apposing membranes together via homophilic adhesion, forming a dense, macroscopic ultrastructure known as the intraperiod line. The C-terminal tail of P0 adheres apposing membranes together in the narrow cytoplasmic compartment of compact myelin, much like myelin basic protein (MBP). In mouse models, the absence of P0, unlike that of MBP or P2, severely disturbs the formation of myelin. Therefore, P0 is the executive molecule of PNS myelin maturation. How and when is P0 trafficked and modified to enable myelin compaction, and how disease mutations that give rise to incurable peripheral neuropathies alter the function of P0, are currently open questions. The potential mechanisms of P0 function in myelination are discussed, providing a foundation for the understanding of mature myelin development and how it derails in peripheral neuropathies. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -
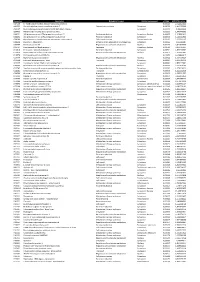
Accession Description Biological Process Cellular Component P-Value SCZ/CTRL Ration A4FU69 EF-Hand Calcium-Binding Domain-Contai
Accession Description Biological Process Cellular component p-Value SCZ/CTRL ration A4FU69 EF-hand calcium-binding domain-containing protein 5 0,001101 2,724427411 A4UGR9 Xin actin-binding repeat-containing protein 2 Cytoskeletal anchoring Cytoplasm 0,006756 1,413953388 A6NCE7 Microtubule-associated proteins 1A/1B light chain 3 beta 2 Cytoplasm 0,001417 1,99612969 B2RPK0 Putative high mobility group protein B1-like 1 0,032314 1,590743352 O00231 26S proteasome non-ATPase regulatory subunit 11 Protein metabolism Cytoplasm; Nucleus 0,000029 1,428664042 O00232 26S proteasome non-ATPase regulatory subunit 12 Protein metabolism Cytoplasm 0,008566 1,544545922 O00264 Membrane-associated progesterone receptor component 1 Cell communication Plasma membrane 0,001459 2,322924147 O00429 Dynamin-1-like protein Mitochondrion organization and biogenesis Cytoplasm 0,006560 0,06391487 O00567 Nucleolar protein 56 Regulation of nucleotide metabolism Nucleus 0,007330 3,842896338 O14737 Programmed cell death protein 5 Apoptosis Cytoplasm; Nucleus 0,006358 3,836727237 O14818 Proteasome subunit alpha type-7 Protein metabolism Cytoplasm 0,030521 1,893928387 O14979 Heterogeneous nuclear ribonucleoprotein D-like Regulation of nucleotide metabolism Nucleus 0,000637 2,150005885 O15078 Centrosomal protein of 290 kDa 0,015359 14,26648619 O15347 High mobility group protein B3 Regulation of nucleotide metabolism Nucleus 0,005500 1,364309014 O15540 Fatty acid-binding protein_ brain Transport Cytoplasm 0,000087 3,125786118 O43237 Cytoplasmic dynein 1 light intermediate -

Symbiotic Adaptations in the Fungal Cultivar of Leaf-Cutting Ants
ARTICLE Received 15 Apr 2014 | Accepted 24 Oct 2014 | Published 1 Dec 2014 DOI: 10.1038/ncomms6675 Symbiotic adaptations in the fungal cultivar of leaf-cutting ants Henrik H. De Fine Licht1,w, Jacobus J. Boomsma2 & Anders Tunlid1 Centuries of artificial selection have dramatically improved the yield of human agriculture; however, strong directional selection also occurs in natural symbiotic interactions. Fungus- growing attine ants cultivate basidiomycete fungi for food. One cultivar lineage has evolved inflated hyphal tips (gongylidia) that grow in bundles called staphylae, to specifically feed the ants. Here we show extensive regulation and molecular signals of adaptive evolution in gene trancripts associated with gongylidia biosynthesis, morphogenesis and enzymatic plant cell wall degradation in the leaf-cutting ant cultivar Leucoagaricus gongylophorus. Comparative analysis of staphylae growth morphology and transcriptome-wide expressional and nucleotide divergence indicate that gongylidia provide leaf-cutting ants with essential amino acids and plant-degrading enzymes, and that they may have done so for 20–25 million years without much evolutionary change. These molecular traits and signatures of selection imply that staphylae are highly advanced coevolutionary organs that play pivotal roles in the mutualism between leaf-cutting ants and their fungal cultivars. 1 Microbial Ecology Group, Department of Biology, Lund University, Ecology Building, SE-223 62 Lund, Sweden. 2 Centre for Social Evolution, Department of Biology, University of Copenhagen, Universitetsparken 15, DK-2100 Copenhagen, Denmark. w Present Address: Section for Organismal Biology, Department of Plant and Environmental Sciences, University of Copenhagen, Thorvaldsensvej 40, DK-1871 Frederiksberg, Denmark. Correspondence and requests for materials should be addressed to H.H.D.F.L.