Influential Parameters of Surface Waters on the Formation of Coating Onto Tio2 Nanoparticles Under Natural Conditions
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Appendix: Supplementary Tables and Raw Data
Appendix: Supplementary tables and raw data Evolutionary genetic analysis of an invasive population of sculpins in the Lower Rhine I n a u g u r a l - D i s s e r t a t i o n zur Erlangung des Doktorgrades der Mathematisch-Naturwissenschaftlichen Fakultät der Universität zu Köln vorgelegt von Arne W. Nolte aus Oldenburg Köln, 2005 Data formats and access: According to the guidelines of the University of Cologne, electronic publication of PhD Theses requires compound documents in PDF–format. On the other hand a simple file format, as for instance ascii-text files, are desirable to have an easy access to datasets. In this appendix, data are formatted as simle text documents and then transformed into PDF format. Thus, one can easily use the “Select Text” option in a PDF viewer (adobe acrobat reader) to copy datasets and paste them into text files. Tables are saved row by row with fields separated by semicolons. Ends of rows are marked by the insertion of “XXX”. Note: In order to recreate a comma separated file (for import into Microsoft Excel) from the texts saved here: 1) all line breaks have to be removed and then 2) the triple XXX has to be replaced by a line break (can be done in a text editor). Otherwise, datasets are composed as indicated in the individual descriptions. Chapter 1 - Supplementary Table 1: Sampled Populations, localities with coordinate data, river basins and references to other studies. Drainage No. Locality GIS References System Volckaert et al. 50°47′N 4°30′ 1 River Neet, S. -

Increasing Importance of Forest Hydrology in a Changing Climate for Forest and Water Management
Increasing Importance of Forest Hydrology in a Changing Climate for Forest and Water Management Prof. Dr. Gebhard Schueler Research Institute for Forest Ecology and Forestry Rhineland-Palatinate Germany Capacity Building Workshop Forest Management and Water Regulation Global Water Partnership – Mediterranean (GWP-Med) 16th – 17th December 2020 Outline Forest Hydrology Landuse Management Ecosystem Services Climate Change Providing Drinking and Process Water Drought – Risk of Forest Health and Vitality Risk Mitigation for Flood and Flashflood Generation and Precautionary Water Retention Forest Hydrology Forest hydrology deals with the water balance of forests and natural woodlands (landscape water balance), in particular, precipitation in and outside of forests (rainfall, snow, mist, interception, throughfall) evapotranspiration, groundwater recharge, runoff behavior (runoff, runoff process) in dependency on forest types (tree species composition, stand type, stand age) and forest management measures. The investigation of the effects of recent forest damage on the water balance has gained special http://www.geodz.com/deu/d/Forsthydrologie Soil Water Reserve in Forests depends upon Precipitation and Evapotranspiration 300 Speyerbach-Schwemmfächer (HA1) 1988-1990 300 / Throughfall Evapotranspiration [l/qm] 275 275 250 250 225 225 200 200 175 175 150 150 125 125 100 100 75 75 plant available soil water reserve [l/qm] reserve soilwater availableplant 50 50 25 25 0 0 01.01.1988 31.01.1988 01.03.1988 31.03.1988 30.04.1988 30.05.1988 29.06.1988 -

Gemeinde Offenbach / Queich
Gemeinde Offenbach / Queich Bebauungsplan „Böhlweg“ Begründung gem. § 9 Abs. 8 BauGB mit Umweltbericht Satzungsfassung Offenbach / Queich Bebauungsplan „Böhlweg“ Satzungsfassung Begründung mit Umweltbericht gem. § 2a BauGB Erstellt im Auftrag der Gemeinde Offenbach / Queich durch BBP Stadtplanung Landschaftsplanung | Kaiserslautern Seite 1 von 39 Offenbach / Queich Bebauungsplan „Böhlweg“ Satzungsfassung Begründung mit Umweltbericht gem. § 2a BauGB INHALTSVERZEICHNIS Ziele, Zwecke und Wesentliche Auswirkungen der Planung gem. § 2 a Nr. 1 BauGB .... 5 A. Erfordernisse und Zielsetzung der Planaufstellung gem. § 1 Abs. 3 BauGB ...... 5 B. Aufstellungsbeschluss ............................................................................................ 5 C. Grundlagen ............................................................................................................... 5 1 Planungsgrundlagen ............................................................................................... 5 2 Lage und Größe des Plangebietes / Grenzen des räumlichen Geltungsbereiches ................................................................................................... 6 3 Bestandssituation (09/2015) .................................................................................... 6 3.1 Nutzung und natürliche Situation ........................................................................... 6 3.2 Schutzgebietsausweisungen ................................................................................. 7 3.3 Geschützte Pflanzen ............................................................................................ -

Wasserversorgungsplan Rheinland-Pfalz Teilgebiet 7
Wasserversorgungsplan Rheinland-Pfalz Karte 4 Teilgebiet 7 - Versorgungsstruktur - Rohwasserförderung und Fremdbezug 2013 STW Deidesheim GmbH (Teilgebiet 6) 245 Haßloch Speyer Re hbach 2 5 h 64 c 69 a Neustadt 1.994 rb a.d. Weinstrasse. 4.659 ye Rhein Spe Haßloch 4 zu 1 Speyer Kirrweiler zu (Pfalz) Sp zu ey 300 Venn- Altdorf 4.396 erb zu ingen 40 5 ach Hanhofen Rodt zu Böb- zu unter ingen zu Dudenhofen Edes- Riet- Maikammer Neustadt Gommers heim burg a.d. Weinstrasse. heim Maikammer Dudenhofen zu zu 3 Kirrweiler Sankt 11 Kirrweiler Edenkoben (Pfalz) zu zu zu Martin (Pfalz) Wilgartswiesen zu zu Zu Gom- Bö- zu Harthausen Römerberg Hainfeld mers- 570 Gommersheim Landau Weyher bin- heim i. d. Pfalz i.d.Pf. zu gen Alt- Flem- zu zu Bö- dorf Edenkoben Altdorf ling- Rosch zu Edes bingen 267 en bach Burrweiler heim Edenkoben Venningen Schwegenheim zu 737 zu zu Frankweiler Rosch- Venningen 6 zu Klingbach bach Wals-- heim Freisbach zu Rhodt unter Großfischlingen Freimersheim Weingarten 470 Albersweiler Böch- 10 zu ingen Rietburg (Pfalz) Gleisweiler Rarnberg zu Weyher Hainfeld (Pfalz) Römerberg zu zu zu i.d.Pfalz Kleinfischlingen Annweiler Siebel- Edes- Hain- 1.071 190 am Trifels dingen heim feld Edesheim zu ch zu zu a 7 Flemlingen nb Westheim Birk - Wals- e heim Burrweiler Rosch- d weiler Flem- o (Pfalz) Lingenfeld Dernbach zu bach Essingen M Trinkwasserverbund Böchingen lingen Knö- 1.025 Eußerthal Gleisweiler ringen Hochstadt Lustadt Böch- Walsheim Lingenfeld Bründelsberg GmbH (Pfalz) 14 ingen Frankweiler 9 Germersheim Rinnthal 790 Zeiskam Queich Wilgartswiesen Albers- 1.822 weiler Landau 600 Annweiler am Siebel- Born- ch heim 24 ei dingen i. -

Heavy Rainfall Provokes Anticoagulant Rodenticides' Release from Baited
Journal Pre-proof Heavy rainfall provokes anticoagulant rodenticides’ release from baited sewer systems and outdoor surfaces into receiving streams Julia Regnery1,*, Robert S. Schulz1, Pia Parrhysius1, Julia Bachtin1, Marvin Brinke1, Sabine Schäfer1, Georg Reifferscheid1, Anton Friesen2 1 Department of Biochemistry, Ecotoxicology, Federal Institute of Hydrology, 56068 Koblenz, Germany 2 Section IV 1.2 Biocides, German Environment Agency, 06813 Dessau-Rosslau, Germany *Corresponding author. Email: [email protected] (J. Regnery); phone: +49 261 1306 5987 Journal Pre-proof A manuscript prepared for possible publication in: Science of the Total Environment May 2020 1 Journal Pre-proof Abstract Prevalent findings of anticoagulant rodenticide (AR) residues in liver tissue of freshwater fish recently emphasized the existence of aquatic exposure pathways. Thus, a comprehensive wastewater treatment plant and surface water monitoring campaign was conducted at two urban catchments in Germany in 2018 and 2019 to investigate potential emission sources of ARs into the aquatic environment. Over several months, the occurrence and fate of all eight ARs authorized in the European Union as well as two pharmaceutical anticoagulants was monitored in a variety of aqueous, solid, and biological environmental matrices during and after widespread sewer baiting with AR-containing bait. As a result, sewer baiting in combined sewer systems, besides outdoor rodent control at the surface, was identified as a substantial contributor of these biocidal active ingredients in the aquatic environment. In conjunction with heavy or prolonged precipitation during bait application in combined sewer systems, a direct link between sewer baiting and AR residues in wastewater treatment plant influent, effluent, and the liver of freshwater fish was established. -

Comprehensive Analysis of the Existing Cross-Border Rail Transport Connections and Missing Links on the Internal EU Borders Final Report
Comprehensive analysis of the existing cross-border rail transport connections and missing links on the internal EU borders Final report Ludger Sippel, Julian Nolte, Simon Maarfield, Dan Wolff, Laure Roux March 2018 EUROPEAN COMMISSION Directorate-General for Regional and Urban Policy Directorate D: European Territorial Cooperation, Macro-regions, Interreg and Programme Implementation I Unit D2: Interreg, Cross-Border Cooperation, Internal Borders Contact: Ana-Paula LAISSY (head of unit), Robert SPISIAK (contract manager) E-mail: [email protected] European Commission B-1049 Brussels EUROPEAN COMMISSION Comprehensive analysis of the existing cross-border rail transport connections and missing links on the internal EU borders Final report Directorate-General for Regional and Urban Policy 2018 EN Europe Direct is a service to help you find answers to your questions about the European Union. Freephone number (*): 00 800 6 7 8 9 10 11 (*) The information given is free, as are most calls (though some operators, phone boxes or hotels may charge you). LEGAL NOTICE The information and views set out in this publication are those of the author(s) and do not necessarily reflect the official opinion of the Commission. The Commission does not guarantee the accuracy of the data included in this study. Neither the Commission nor any person acting on the Commission’s behalf may be held responsible for the use which may be made of the information contained therein. More information on the European Union is available on the Internet (http://www.europa.eu). Luxembourg: Publications Office of the European Union, 2018 ISBN 978-92-79-85821-5 doi: 10.2776/69337 © European Union, 2018 Reproduction is authorised provided the source is acknowledged. -

Transport Operators • Marketing Strategies for Ridership and Maximising Revenue • LRT Solutions for Mid-Size Towns
THE INTERNATIONAL LIGHT RAIL MAGAZINE www.lrta.org www.tautonline.com FEBRUARY 2015 NO. 926 NEW TRAMWAYS: CHINA EMBRACES LIGHT RAIL Houston: Two new lines for 2015 and more to come Atlanta Streetcar launches service Paris opens T6 and T8 tramlines Honolulu metro begins tracklaying ISSN 1460-8324 UK devolution San Diego 02 £4.25 Will regional powers Siemens and partners benefit light rail? set new World Record 9 771460 832043 2015 INTEGRATION AND GLOBALISATION Topics and panel debates include: Funding models and financial considerations for LRT projects • Improving light rail's appeal and visibility • Low Impact Light Rail • Harnessing local suppliers • Making light rail procurement more economical • Utilities replacement and renewal • EMC approvals and standards • Off-wire and energy recovery systems • How can regional devolution benefit UK LRT? • Light rail safety and security • Optimising light rail and traffic interfaces • Track replacement & renewal – Issues and lessons learned • Removing obsolescence: Modernising the UK's second-generation systems • Revenue protection strategies • Big data: Opportunities for transport operators • Marketing strategies for ridership and maximising revenue • LRT solutions for mid-size towns Do you have a topic you’d like to share with a forum of 300 senior decision-makers? We are still welcoming abstracts for consideration until 12 January 2015. June 17-18, 2015: Nottingham, UK Book now via www.tautonline.com SUPPORTED BY 72 CONTENTS The official journal of the Light Rail Transit Association FEBRUARY 2015 Vol. 78 No. 926 www.tramnews.net EDITORIAL 52 EDITOR Simon Johnston Tel: +44 (0)1733 367601 E-mail: [email protected] 13 Orton Enterprise Centre, Bakewell Road, Peterborough PE2 6XU, UK ASSOCIATE EDITOR Tony Streeter E-mail: [email protected] WORLDWIDE EDITOR Michael Taplin Flat 1, 10 Hope Road, Shanklin, Isle of Wight PO37 6EA, UK. -

Annales Scientifiques Wissenschaftliches Jahrbuch 2011
2011 2012 SOMMAIRE TOME / BAND 16 – 2011-2012 • Première observation en France de l’Ecrevisse calicot, Orconnectes immunis (Hagen, 1870) - COLLAS M.,BEINSTEINER D., FRITSCH S., MORELLE S. & L’HOSPITALIER M. ............................................................ 18-36 • Die Forsthäuser in und um Speyerbrunn Baukulturelles Erbe und Symbol für die Kulturlandschaft Pfälzerwald - FINKBEINER J. .................. 38-73 • Flusskrebse im Einzugsgebiet von Saarbach und Eppenbrunner Bach - Er- fassung und grenzüberschreitender Schutz autochthoner Flusskrebsarten im Biosphärenreservat „Pfälzerwald – Vosges du Nord“ - IDELBERGER S., SCHLEICH S., OTT J. & WAGNER M. ....................................... 74-98 • Wooge auf die Agenda des Biosphärenreservats ? Bedeutung, Bewertung und zukünftige Bewirtschaftung der prägenden Gewässer im Pfälzerwald - KOEHLER G., FREY W., HAUPTLORENZ H. & SCHINDLER H. ..... 100-117 • Der Biosphärenturm - ein innovatives Alleinstellungsmerkmal zur Baum- kronenforschung - LAKATOS M., WIRTH R., SPITZLEY P., LEDERER F. & BÜDEL B. ............................................................................... 118-129 • Suivi de la mortalité routière de la faune le long de la route départementale reliant Bitche à Sarreguemines - MORELLE S. & GENOT J.-C. ....... 130-143 Annales • La conservation des arbres d’intérêt biologique dans le Parc naturel régional des Vosges du Nord. Un premier bilan - PASCAL B. .................... 144-153 scientifiques • La réactualisation des ZNIEFF dans le Parc naturel régional des Vosges -
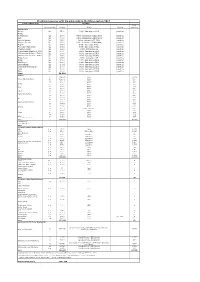
Stocking Measures with Big Salmonids in the Rhine System 2017
Stocking measures with big salmonids in the Rhine system 2017 Country/Water body Stocking smolt Kind and stage Number Origin Marking equivalent Switzerland Wiese Lp 3500 Petite Camargue B1K3 genetics Rhine Riehenteich Lp 1.000 Petite Camargue K1K2K4K4a genetics Birs Lp 4.000 Petite Camargue K1K2K4K4a genetics Arisdörferbach Lp 1.500 Petite Camargue F1 Wild genetics Hintere Frenke Lp 2.500 Petite Camargue K1K2K4K4a genetics Ergolz Lp 3.500 Petite Camargue K7C1 genetics Fluebach Harbotswil Lp 1.300 Petite Camargue K7C1 genetics Magdenerbach Lp 3.900 Petite Camargue K5 genetics Möhlinbach (Bachtele, Möhlin) Lp 600 Petite Camargue B7B8 genetics Möhlinbach (Möhlin / Zeiningen) Lp 2.000 Petite Camargue B7B8 genetics Möhlinbach (Zuzgen, Hellikon) Lp 3.500 Petite Camargue B7B8 genetics Etzgerbach Lp 4.500 Petite Camargue K5 genetics Rhine Lp 1.000 Petite Camargue B2K6 genetics Old Rhine Lp 2.500 Petite Camargue B2K6 genetics Bachtalbach Lp 1.000 Petite Camargue B2K6 genetics Inland canal Klingnau Lp 1.000 Petite Camargue B2K6 genetics Surb Lp 1.000 Petite Camargue B2K6 genetics Bünz Lp 1.000 Petite Camargue B2K6 genetics Sum 39.300 France L0 269.147 Allier 13457 Rhein (Alt-/Restrhein) L0 142.000 Rhine 7100 La 31.500 Rhine 3150 L0 5.000 Rhine 250 Doller La 21.900 Rhine 2190 L0 2.500 Rhine 125 Thur La 12.000 Rhine 1200 L0 2.500 Rhine 125 Lauch La 5.000 Rhine 500 Fecht und Zuflüsse L0 10.000 Rhine 500 La 39.000 Rhine 3900 L0 4.200 Rhine 210 Ill La 17.500 Rhine 1750 Giessen und Zuflüsse L0 10.000 Rhine 500 La 28.472 Rhine 2847 L0 10.500 Rhine 525 -

Joint Meeting of Working Groups 1, 3 and 5 of INLA
Joint Meeting of Working Groups 1, 3 and 5 of INLA WHEN: 03-04 May 2017 WHERE: Karlsruhe - Germany Organizer: International Nuclear Law Association We are pleased to announce that the next Joint Meeting of Working Group 1 (Safety & Regulation), Working Group 3 (New Build) and Working Group 5 (Waste Management) of INLA will take place on Wednesday, 3rd May and Thursday, 4th May 2017 at the Joint Research Centre of the European Commission in Karlsruhe. REGISTRATION Please report your availability for attendance to Ms Alexandra Van Kalleveen (Alexandra.VAN- [email protected]) before end of March 2017. Participation is free of charge. VENUE European Commission Joint Research Centre (JRC) Directorate G - Nuclear Safety and Security JRC Karlsruhe Site Hermann-von-Helmholtz-Platz 1 76344 Eggenstein-Leopoldshafen Germany SECURITY POLICY Everyone requesting access to the site must be registered in advance therefore registration forms will be sent to you upon confirming your participation. Also a copy of identity card or passport has to be provided. Nuclear installations are subject to strict security rules. In this context, it is prohibited to take pictures or movies within the premises of the JRC Karlsruhe unless an explicit derogation has been delivered. In order to implement this requirement, visitors entering the site are, as a general rule, not allowed to bring with them any mobile phones with cameras or other technical equipment for taking pictures or movies (laptops included). If, nevertheless, this is absolutely necessary you should fill in the form indicating the reason why the devices are needed. Please indicate this to the meeting secretary ([email protected]). -

812402.En Pe 440.438
WRITTEN QUESTION E-2642/10 by Franziska Katharina Brantner (Verts/ALE) to the Commission Subject: Flood retention basins for Waldsee/Altrip/Neuhofen; implementation of EU directive The Land of Rhineland-Palatinate is planning to build two flood retention basins in the municipalities of Waldsee, Altrip and Neuhofen in the Rhein-Pfalz district: the Waldsee/Altrip/Neuhofen flood retention basin (W/A/N polder) and the Rehbach polders on the road between Altrip and Rheingönheim (K7). The proposed construction works will directly affect the conservation objectives of Altrip's inhabitants, the district's protected areas – in which some of the construction work will be carried out – and the nearby bodies of surface water and drainage ditches. The planned flood retention basins will encroach on parts of several flora and fauna habitats and bird protection areas. Even now, when the Rhine floods, the groundwater in areas which are prone to waterlogging rises to the surface, and this is the case in the inhabited area of Altrip. When the W/A/N polder is flooded – both the 'controlled' and the 'uncontrolled' parts – it is likely that the pressurised water and groundwater problem will worsen. Moreover, according to the planning authority (SGD Süd), when the W/A/N polder is flooded the K13 road to Waldsee is left 20 centimetres under water, cutting off one emergency evacuation route for the inhabitants of Altrip. The only remaining evacuation route would then be the K7 to Rheingönheim on the old main dyke of the Rhine, which will then be weakened from both sides when the Rhine floods. -

CE/CL/Annex IV/En 1 ANNEX IV AGREEMENT on SANITARY AND
549 der Beilagen XXII. GP - Beschluss NR - Englische Anhänge 4 (Normativer Teil) 1 von 825 ANNEX IV AGREEMENT ON SANITARY AND PHYTOSANITARY MEASURES APPLICABLE TO TRADE IN ANIMALS AND ANIMAL PRODUCTS, PLANTS, PLANT PRODUCTS AND OTHER GOODS AND ANIMAL WELFARE (Referred to in Article 89(2) of the Association Agreement) THE PARTIES, as defined in Article 197 of the Association Agreement: DESIRING to facilitate trade between the Community and Chile in animals and animal products, plants, plant products and other goods, whilst safeguarding public, animal and plant health; CONSIDERING that the implementation of this Agreement is to take place in accordance with the internal procedures and legislative processes of the Parties; CONSIDERING that recognition of equivalence will be gradual and progressive and should apply to priority sectors; CE/CL/Annex IV/en 1 2 von 825 549 der Beilagen XXII. GP - Beschluss NR - Englische Anhänge 4 (Normativer Teil) CONSIDERING that one of the objectives of Part IV, Title I of the Association Agreement is to liberalise trade in goods in accordance with the GATT 1994 progressively and reciprocally; REAFFIRMING their rights and obligations under the WTO Agreement and its Annexes and in particular the SPS Agreement; DESIRING to ensure full transparency as regards sanitary, phytosanitary measures applicable to trade, to have a common understanding of the WTO SPS Agreement and to implement its principles and provisions; RESOLVED to take the fullest account of the risk of spread of animal infections, diseases and pests and of the measures put in place to control and eradicate such infections, diseases and pests, to protect public, animal and plant health while avoiding unnecessary disruptions to trade; WHEREAS, given the importance of animal welfare, with the aim of developing animal welfare standards and given its relation with veterinary matters, it is appropriate to include this issue in this Agreement and to examine animal welfare standards taking into account the development in the competent international standards organisations.