A Protease Enzyme Preparation from a Recombinant Strain of Trichoderma Reesei
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
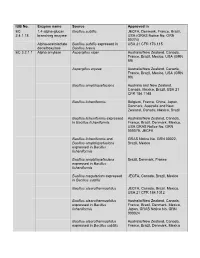
IUB No. Enzyme Name Source Approved in EC 1,4-Alpha-Glucan Bacillus Subtilis JECFA, Denmark, France, Brazil, 2.4.1.18 Branching Enzyme USA (GRAS Notice No
IUB No. Enzyme name Source Approved in EC 1,4-alpha-glucan Bacillus subtilis JECFA, Denmark, France, Brazil, 2.4.1.18 branching enzyme USA (GRAS Notice No. GRN 00274) Alpha-acetolactate Bacillus subtilis expressed in USA 21 CFR 173.115 decarboxylase Bacillus brevis EC 3.2.1.1 Alpha amylase Aspergillus niger Australia/New Zealand, Canada, France, Brazil, Mexico, USA (GRN 89) Aspergillus oryzae Australia/New Zealand, Canada, France, Brazil, Mexico, USA (GRN 90) Bacillus amyloliquefaciens Australia and New Zealand, Canada, Mexico, Brazil, USA 21 CFR 184.1148 Bacillus licheniformis Belgium, France, China, Japan, Denmark, Australia and New Zealand, Canada, Mexico, Brazil Bacillus licheniformis expressed Australia/New Zealand, Canada, in Bacillus licheniformis France, Brazil, Denmark, Mexico, USA GRAS Notice No. GRN 000079, JECFA Bacillus licheniformis and GRAS Notice No. GRN 00022, Bacillus amyloliquefaciens Brazil, Mexico expressed in Bacillus licheniformis Bacillus amyloliquefaciens Brazil, Denmark, France expressed in Bacillus licheniformis Bacillus megaterium expressed JECFA, Canada, Brazil, Mexico in Bacillus subtilis Bacillus stearothermophilus JECFA, Canada, Brazil, Mexico, USA 21 CFR 184.1012 Bacillus stearothermophilus Australia/New Zealand, Canada, expressed in Bacillus France, Brazil, Denmark, Mexico, licheniformis Japan, GRAS Notice No. GRN 000024 Bacillus stearothermophilus Australia/New Zealand, Canada, expressed in Bacillus subtilis France, Brazil, Denmark, Mexico Bacillus stearothermophilus Australia/New Zealand, Canada, (Geobacillus France, Europa, Brazil, Mexico, stearothermophilus) USA (GRN 594) Bacillus subtilis Australia and New Zealand, Canada, Mexico, Brazil, USA 21 CFR 184.1148 Rhizopus delemar Brazil Rhizopus oryzae Brazil, Mexico, USA GRAS Notice No. GRN 000090 Thermoccocales expressed in Brazil, USA (GRN126) Pseudomonas fluorecens Rhizomucor pusillus expressed Brazil, Denmark, France, Mexico, in Aspergillus niger Australia and New Zealand. -

(12) Patent Application Publication (10) Pub. No.: US 2016/0346364 A1 BRUNS Et Al
US 2016.0346364A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2016/0346364 A1 BRUNS et al. (43) Pub. Date: Dec. 1, 2016 (54) MEDICAMENT AND METHOD FOR (52) U.S. Cl. TREATING INNATE IMMUNE RESPONSE CPC ........... A61K 38/488 (2013.01); A61K 38/482 DISEASES (2013.01); C12Y 304/23019 (2013.01); C12Y 304/21026 (2013.01); C12Y 304/23018 (71) Applicant: DSM IPASSETS B.V., Heerlen (NL) (2013.01); A61K 9/0053 (2013.01); C12N 9/62 (2013.01); A23L 29/06 (2016.08); A2ID 8/042 (72) Inventors: Maaike Johanna BRUINS, Kaiseraugst (2013.01); A23L 5/25 (2016.08); A23V (CH); Luppo EDENS, Kaiseraugst 2002/00 (2013.01) (CH); Lenneke NAN, Kaiseraugst (CH) (57) ABSTRACT (21) Appl. No.: 15/101,630 This invention relates to a medicament or a dietary Supple (22) PCT Filed: Dec. 11, 2014 ment comprising the Aspergillus niger aspergilloglutamic peptidase that is capable of hydrolyzing plant food allergens, (86). PCT No.: PCT/EP2014/077355 and more particularly, alpha-amylase/trypsin inhibitors, thereby treating diseases due to an innate immune response S 371 (c)(1), in humans, and/or allowing to delay the onset of said (2) Date: Jun. 3, 2016 diseases. The present invention relates to the discovery that (30) Foreign Application Priority Data the Aspergillus niger aspergilloglutamic peptidase is capable of hydrolyzing alpha-amylase/trypsin inhibitors that are Dec. 11, 2013 (EP) .................................. 13196580.8 present in wheat and related cereals said inhibitors being strong inducers of innate immune response. Furthermore, Publication Classification the present invention relates to a method for hydrolyzing alpha-amylase/trypsin inhibitors comprising incubating a (51) Int. -

Evaluation of Anthelmintic Activity of Carica Papaya Latex Using Pheritima Posthuma
Research Article Vol 2/Issue 1/Jan-Mar 2012 EVALUATION OF ANTHELMINTIC ACTIVITY OF CARICA PAPAYA LATEX USING PHERITIMA POSTHUMA LAKSHMI KANTA KANTHAL1*, PRASENJIT MONDAL2, SOMNATH DE4, SOMA JANA3, S. ANEELA4 AND K. SATYAVATHI1 1Koringa college of Pharmacy, Korangi, Tallarevu (M), East Godavari Dist., A.P. 2Vaageswari College Of Pharmacy, Karimnagar, A.P. 3Vaageswari Institute Of Pharmaceutical sciences, Karimnagar, A.P 4Dr.Samuel George Institute Of Pharmaceutical Sciences, Markapur, A.P. ABSTRACT The aim of present study is to evaluate Anthelmintic potential of latex of Carica papaya using Pheretima posthuma as test worms. Various concentrations (100%, 50%, and 20%) of Carica papaya latex were tested in the assay, which involved determination of time of paralysis (P) and time of death (D) of the worms. It show shortest time of paralysis (P=24.5 min) and death (D=56min) in 100% concentration, while the time of paralysis and death will increase in 50% concentration (P=28min&D=64min) and in 20% concentration (P=34min&D=74min) respectively as compare to Piperazine citrate (10mg/ml) used as standard reference (P= 24 min& D= 54) and distilled water as control. The results of present study indicated that the latex of Carica papaya showed significantly demonstrated paralysis, and also caused death of worms especially at higher concentration as compared to standard reference Piperazine citrate and control.From the result it is conclude that the latex of Carica papaya showed significant Anthelmintic activity. Key words : Pheretima posthuma, Anthelmintic, Carica papaya latex, Piperazine citrate. 1. INTRODUCTION Helminthiasis is a disease in which a part of the .2005).The papaya is a short-lived, fast-growing, body is infested with worms such as pinworm, woody, large herb to 10 or 12 feet in height. -

Progress in the Field of Aspartic Proteinases in Cheese Manufacturing
Progress in the field of aspartic proteinases in cheese manufacturing: structures, functions, catalytic mechanism, inhibition, and engineering Sirma Yegin, Peter Dekker To cite this version: Sirma Yegin, Peter Dekker. Progress in the field of aspartic proteinases in cheese manufacturing: structures, functions, catalytic mechanism, inhibition, and engineering. Dairy Science & Technology, EDP sciences/Springer, 2013, 93 (6), pp.565-594. 10.1007/s13594-013-0137-2. hal-01201447 HAL Id: hal-01201447 https://hal.archives-ouvertes.fr/hal-01201447 Submitted on 17 Sep 2015 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Dairy Sci. & Technol. (2013) 93:565–594 DOI 10.1007/s13594-013-0137-2 REVIEW PAPER Progress in the field of aspartic proteinases in cheese manufacturing: structures, functions, catalytic mechanism, inhibition, and engineering Sirma Yegin & Peter Dekker Received: 25 February 2013 /Revised: 16 May 2013 /Accepted: 21 May 2013 / Published online: 27 June 2013 # INRA and Springer-Verlag France 2013 Abstract Aspartic proteinases are an important class of proteinases which are widely used as milk-coagulating agents in industrial cheese production. They are available from a wide range of sources including mammals, plants, and microorganisms. -

Serine Proteases with Altered Sensitivity to Activity-Modulating
(19) & (11) EP 2 045 321 A2 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 08.04.2009 Bulletin 2009/15 C12N 9/00 (2006.01) C12N 15/00 (2006.01) C12Q 1/37 (2006.01) (21) Application number: 09150549.5 (22) Date of filing: 26.05.2006 (84) Designated Contracting States: • Haupts, Ulrich AT BE BG CH CY CZ DE DK EE ES FI FR GB GR 51519 Odenthal (DE) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • Coco, Wayne SK TR 50737 Köln (DE) •Tebbe, Jan (30) Priority: 27.05.2005 EP 05104543 50733 Köln (DE) • Votsmeier, Christian (62) Document number(s) of the earlier application(s) in 50259 Pulheim (DE) accordance with Art. 76 EPC: • Scheidig, Andreas 06763303.2 / 1 883 696 50823 Köln (DE) (71) Applicant: Direvo Biotech AG (74) Representative: von Kreisler Selting Werner 50829 Köln (DE) Patentanwälte P.O. Box 10 22 41 (72) Inventors: 50462 Köln (DE) • Koltermann, André 82057 Icking (DE) Remarks: • Kettling, Ulrich This application was filed on 14-01-2009 as a 81477 München (DE) divisional application to the application mentioned under INID code 62. (54) Serine proteases with altered sensitivity to activity-modulating substances (57) The present invention provides variants of ser- screening of the library in the presence of one or several ine proteases of the S1 class with altered sensitivity to activity-modulating substances, selection of variants with one or more activity-modulating substances. A method altered sensitivity to one or several activity-modulating for the generation of such proteases is disclosed, com- substances and isolation of those polynucleotide se- prising the provision of a protease library encoding poly- quences that encode for the selected variants. -

Chymopapain Chemonucleolysis: CT Changes After Treatment
321 Chymopapain Chemonucleolysis: CT Changes after Treatment Lindell R. Gentry 1. 2 Chymopapain chemonucleolysis is now used extensively in this country to treat lumbar Patrick A. Turski1 disk herniation. Despite increasing experience in patient selection, there continue to be Charles M. Strother1 patients who do not respond to treatment and require diagnostic reevaluation. Interpre Manucher J. Javid3 tation of postchemonucleolysis computed tomographic (CT) scans in these patients Joseph F. Sackett 1 requires a knowledge of the CT changes that normally occur after treatment with chemonucleolysis. To define these temporal changes, a prospective CT evaluation was performed of 29 treated interspaces in 26 patients who returned for routine postche monucleolysis follOW-Up. Despite a successful clinical response in 17 of 21 patients, changes in the size, location, shape, homogeneity, and density of the disk herniation were uncommon at the 6 week follow-up. In 24 treated interspaces, the most common changes at 6 week CT follow-up were the development of vacuum phenomenon in three (12.5%) and a slight decrease in the size of two (8.3%) disk herniations. A successful response was noted in 17 of 21 patients scanned at 6 month follow-up, with five (22.7%) of 22 injected interspaces exhibiting vacuum phenomenon and 13 (59.1%) interspaces showing an observable decrease in the size of the disk herniation. Early improvement of sciatica after chemonucleolysis often occurs without a change in the size of the disk herniation and may be mediated by chymopapain-induced disk-space narrowing. Con tinued improvement may be accompanied by both a decrease in the disk height and a reduction in the size of the disk protrusion. -

Collection of Information on Enzymes a Great Deal of Additional Information on the European Union Is Available on the Internet
European Commission Collection of information on enzymes A great deal of additional information on the European Union is available on the Internet. It can be accessed through the Europa server (http://europa.eu.int). Luxembourg: Office for Official Publications of the European Communities, 2002 ISBN 92-894-4218-2 © European Communities, 2002 Reproduction is authorised provided the source is acknowledged. Final Report „Collection of Information on Enzymes“ Contract No B4-3040/2000/278245/MAR/E2 in co-operation between the Federal Environment Agency Austria Spittelauer Lände 5, A-1090 Vienna, http://www.ubavie.gv.at and the Inter-University Research Center for Technology, Work and Culture (IFF/IFZ) Schlögelgasse 2, A-8010 Graz, http://www.ifz.tu-graz.ac.at PROJECT TEAM (VIENNA / GRAZ) Werner Aberer c Maria Hahn a Manfred Klade b Uli Seebacher b Armin Spök (Co-ordinator Graz) b Karoline Wallner a Helmut Witzani (Co-ordinator Vienna) a a Austrian Federal Environmental Agency (UBA), Vienna b Inter-University Research Center for Technology, Work, and Culture - IFF/IFZ, Graz c University of Graz, Department of Dermatology, Division of Environmental Dermatology, Graz Executive Summary 5 EXECUTIVE SUMMARY Technical Aspects of Enzymes (Chapter 3) Application of enzymes (Section 3.2) Enzymes are applied in various areas of application, the most important ones are technical use, manufacturing of food and feedstuff, cosmetics, medicinal products and as tools for re- search and development. Enzymatic processes - usually carried out under mild conditions - are often replacing steps in traditional chemical processes which were carried out under harsh industrial environments (temperature, pressures, pH, chemicals). Technical enzymes are applied in detergents, for pulp and paper applications, in textile manufacturing, leather industry, for fuel production and for the production of pharmaceuticals and chiral substances in the chemical industry. -

Penicillopepsin-JT2, a Recombinant Enzyme from Penicillium Janthinellum and the Contribution of a Hydrogen Bond in Subsite S3 to Kcat
Protein Science ~2000!, 9:991–1001. Cambridge University Press. Printed in the USA. Copyright © 2000 The Protein Society Penicillopepsin-JT2, a recombinant enzyme from Penicillium janthinellum and the contribution of a hydrogen bond in subsite S3 to kcat QING-NA CAO,1,3 MARLENE STUBBS,1 KENNY Q.P. NGO,1 MICHAEL WARD,2 ANNIE CUNNINGHAM,1 EMIL F. PAI,1 GUANG-CHOU TU,1,3 and THEO HOFMANN1 1 Department of Biochemistry, University of Toronto, Toronto, Ontario M5S 1A8, Canada 2 Genencor International, Inc., 925 Page Mill Road, Palo Alto, California 94304-1013 ~Received August 30, 1999; Final Revision February 7, 2000; Accepted March 10, 2000! Abstract The nucleotide sequence of the gene ~ pepA! of a zymogen of an aspartic proteinase from Penicillium janthinellum with a 71% identity in the deduced amino acid sequence to penicillopepsin ~which we propose to call penicillopepsin-JT1! has been determined. The gene consists of 60 codons for a putative leader sequence of 20 amino acid residues, a sequence of about 150 nucleotides that probably codes for an activation peptide and a sequence with two introns that codes for the active aspartic proteinase. This gene, inserted into the expression vector pGPT-pyrG1, was expressed in an aspartic proteinase-free strain of Aspergillus niger var. awamori in high yield as a glycosylated form of the active enzyme that we call penicillopepsin-JT2. After removal of the carbohydrate component with endoglycosidase H, its relative molecular mass is between 33,700 and 34,000. Its kinetic properties, especially the rate-enhancing effects of the presence of alanine residues in positions P3 and P29 of substrates, are similar to those of penicillopepsin-JT1, endothia- pepsin, rhizopuspepsin, and pig pepsin. -

Review Article the Role of Microbial Aspartic Protease Enzyme in Food and Beverage Industries
Hindawi Journal of Food Quality Volume 2018, Article ID 7957269, 15 pages https://doi.org/10.1155/2018/7957269 Review Article The Role of Microbial Aspartic Protease Enzyme in Food and Beverage Industries Jermen Mamo and Fassil Assefa Microbial, Cellular and Molecular Biology Department, College of Natural Science, Addis Ababa University, P.O. Box 1176, Addis Ababa, Ethiopia Correspondence should be addressed to Jermen Mamo; [email protected] Received 3 April 2018; Revised 16 May 2018; Accepted 29 May 2018; Published 3 July 2018 Academic Editor: Antimo Di Maro Copyright © 2018 Jermen Mamo and Fassil Assefa. is is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Proteases represent one of the three largest groups of industrial enzymes and account for about 60% of the total global enzymes sale. According to the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology, proteases are classied in enzymes of class 3, the hydrolases, and the subclass 3.4, the peptide hydrolases or peptidase. Proteases are generally grouped into two main classes based on their site of action, that is, exopeptidases and endopeptidases. Protease has also been grouped into four classes based on their catalytic action: aspartic, cysteine, metallo, and serine proteases. However, lately, three new systems have been dened: the threonine-based proteasome system, the glutamate-glutamine system of eqolisin, and the serine-glutamate-aspartate system of sedolisin. Aspartic proteases (EC 3.4.23) are peptidases that display various activities and specicities. -

Treatment of Cashew Extracts with Aspergillopepsin Reduces Ige Binding to Cashew Allergens
Journal of Applied Biology & Biotechnology Vol. 4 (02), pp. 001-010, March-April, 2016 Available online at http://www.jabonline.in DOI: 10.7324/JABB.2016.40201 Treatment of cashew extracts with Aspergillopepsin reduces IgE binding to cashew allergens Cecily B. DeFreece1, Jeffrey W. Cary2, Casey C. Grimm2, Richard L. Wasserman3, Christopher P Mattison2* 1Department of Biology, Xavier University of Louisiana, New Orleans, LA, USA. 2USDA-ARS, Southern Regional Research Center, New Orleans, LA, USA. 3Allergy Partners of North Texas Research, Department of Pediatrics, Medical City Children’s Hospital, Dallas, Texas, USA. ARTICLE INFO ABSTRACT Article history: Enzymes from Aspergillus fungal species are used in many industrial and pharmaceutical applications. Received on: 21/10/2015 Aspergillus niger and Aspergillus oryzae were cultured on media containing cashew nut flour to identify secreted Revised on: 06/12/2015 proteins that may be useful as future food allergen processing enzymes. Mass-spectrometric analysis of secreted Accepted on: 20/12/2015 proteins and protein bands from SDS-PAGE gels indicated the presence of at least 63 proteins. The majority of Available online: 21/04/2016 these proteins were involved in carbohydrate metabolism, but there were also enzymes involved in lipid and protein metabolism. It is likely that some of these enzymes are specifically upregulated in response to cashew Key words: nut protein, and study of these enzymes could aid our understanding of cashew nut metabolism. Aspergillus, cashew, food Aspergillopepsin from A. niger was one of the proteolytic enzymes identified, and 6 distinct peptides were allergy, immunoglobulin E, matched to this protein providing 22% coverage of the protein. -

DUAL ROLE of CATHEPSIN D: LIGAND and PROTEASE Martin Fuseka, Václav Větvičkab
Biomed. Papers 149(1), 43–50 (2005) 43 © M. Fusek, V. Větvička DUAL ROLE OF CATHEPSIN D: LIGAND AND PROTEASE Martin Fuseka, Václav Větvičkab* a Institute of Organic Chemistry and Biochemistry, CAS, Prague, Czech Republic, and b University of Louisville, Department of Pathology, Louisville, KY40292, USA, e-mail: [email protected] Received: April 15, 2005; Accepted (with revisions): June 20, 2005 Key words: Cathepsin D/Procathepsin D/Cancer/Activation peptide/Mitogenic activity/Proliferation Cathepsin D is peptidase belonging to the family of aspartic peptidases. Its mostly described function is intracel- lular catabolism in lysosomal compartments, other physiological effect include hormone and antigen processing. For almost two decades, there have been an increasing number of data describing additional roles imparted by cathepsin D and its pro-enzyme, resulting in cathepsin D being a specific biomarker of some diseases. These roles in pathological conditions, namely elevated levels in certain tumor tissues, seem to be connected to another, yet not fully understood functionality. However, despite numerous studies, the mechanisms of cathepsin D and its precursor’s actions are still not completely understood. From results discussed in this article it might be concluded that cathepsin D in its zymogen status has additional function, which is rather dependent on a “ligand-like” function then on proteolytic activity. CATHEPSIN D – MEMBER PRIMARY, SECONDARY AND TERTIARY OF ASPARTIC PEPTIDASES FAMILY STRUCTURES OF ASPARTIC PEPTIDASES Major function of cathepsin D is the digestion of There is a high degree of sequence similarity among proteins and peptides within the acidic compartment eukaryotic members of the family of aspartic peptidases, of lysosome1. -

Investigating the Impact of Mpapr1, an Aspartic Protease from the Yeast Metschnikowia Pulcherrima, on Wine Properties
THÈSE EN COTUTELLE PRÉSENTÉE POUR OBTENIR LE GRADE DE DOCTEUR DE L’UNIVERSITÉ DE BORDEAUX ET DE L’UNIVERSITÉ DE STELLENBOSCH ÉCOLE DOCTORALE DES SCIENCES DE LA VIE ET DE LA SANTÉ SPÉCIALITÉ ŒNOLOGIE FACULTY OF AGRISCIENCES Par Louwrens THERON ETUDE DE L’IMPACT DE MPAPR1, UNE PROTEASE ASPARTIQUE DE LA LEVURE METSCHNIKOWIA PULCHERRIMA, SUR LES PROPRIETES DU VIN Sous la direction de Benoit DIVOL et de Marina BELY Soutenue le 27 janvier 2017 Membres du jury: Mme. LE HENAFF-LE MARREC Claire, Professeur à l’université de Bordeaux Président M. MARANGON Matteo, Chargé de recherche à l’université de Padoue Rapporteur Mme. CAMARASA Carole, Chargée de recherche à l’INRA de Montpellier Rapporteur M. BAUER Florian, Professeur à l’université de Stellenbosch Examinateur Titre : Etude de l’impact de MpAPr1, une protéase aspartique de la levure Metschnikowia pulcherrima, sur les propriétés du vin Résumé : L'élimination des protéines est une étape clé lors de la production du vin blanc afin d'éviter l'apparition éventuelle d'un voile inoffensif mais inesthétique. Des solutions de rechange à l'utilisation de la bentonite sont activement recherchées en raison des problèmes technologiques, organoleptiques et de durabilité associés à son utilisation. Dans cette étude, MpAPr1, une protéase aspartique extracellulaire préalablement isolée et partiellement caractérisée à partir de la levure Metschnikowia pulcherrima, a été clonée et exprimée de manière hétérologue dans la levure Komagataella pastoris. Les propriétés enzymatiques de MpAPr1 ont été initialement caractérisées dans un extrait brut. Après plusieurs essais faisant appel à différentes techniques, MpAPr1 a été purifié avec succès par chromatographie échangeusede cations.